Nucleic Acid Quantification
February 2019
The quantification of nucleic acids is a necessary procedure after isolation from samples such as tissues, cells or body fluids. Downstream applications include PCR, RT-PCR, sequencing, restriction digestions and ligations. All these applications involve enzymatic reactions where efficiency is dependent on the relative concentrations of nucleic acid, enzyme and other reactants, hence the need for quantification. Amounts of nucleic acid isolated from most kits can range anywhere from ng to µg and are typically eluted in 10 to 100 µL volumes. Nucleic acid concentrations can range from sub ng/µL to thousands of ng/µL. Spectrophotometry using the intrinsic chromophores within the purine and pyrimidine bases is a very popular method for nucleic acid quantification as it is a simple, accurate and non-destructive method for the measurement of nucleic acid over much of the range of concentrations described above; however some samples require additional sensitivity to quantify the low levels of nucleic acid present. These samples rely on the use of fluorogenic dyes that when bound to the nucleic acids, become highly fluorescent.
Traditionally, nucleic acid detection has been performed in standard 1 cm path length cuvettes, but dilution of the sample is typically required to provide sufficient volume for the cuvette measurement or to avoid Beer's Law non-linearity issues at high optical density. This can be remedied by the use of micro-volume analysis where sample size requirements are only a small fraction of the collected sample and as the path length is typically 20 times smaller than a cuvette, the samples can be directly analyzed. Finally, for those labs with a need for analyzing many samples, the use of microplates can drastically improve sample throughput.
This edition of TekTalk will cover the many aspects of nucleic acid quantification and focus on BioTek's products that enable these measurements.
Featured Applications
Analytical Performance of Nucleic Acid Micro-Volume Quantification Using the Epoch Spectrophotometer System
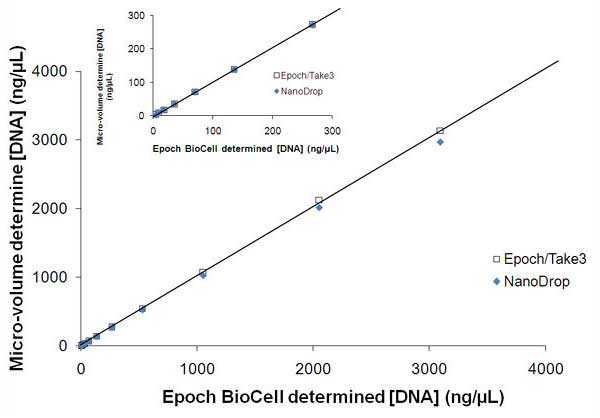
Nucleic acids such as DNA and RNA are derived from a variety of biological sources for potential use in downstream applications such as sequencing and gene expression analysis. Here we demonstrate the analytical performance of the Epoch Spectrophotometer System and especially the Take3 Multi-Volume Plate with regard to linear dynamic range, limit of detection, precision and accuracy using micro-volume samples to verify its broad applicability to nucleic acid quantification. Comparative data was derived on Epoch Spectrophotometer System and NanoDrop: performance was shown to be equivalent.
Quantification of dsDNA using Qubit® dsDNA Broad Range (BR) Assay Kit
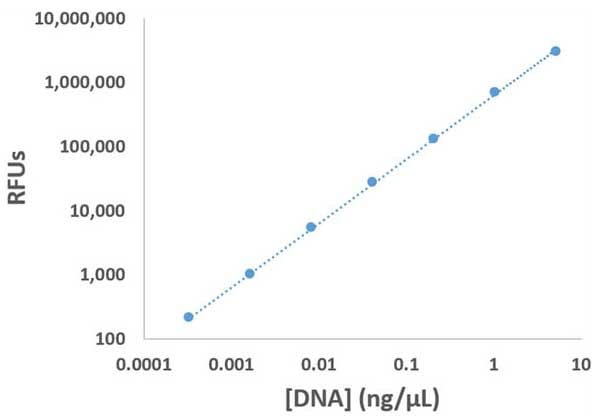
Nucleic acid quantitation remains a central component of myriad workflows across a wide spectrum of biological sciences. Many of these workflows require accurate determination of sample concentration from very small sample volumes. The use of enhancing reagents provides a means to perform such analyses while conserving precious sample.
Application Guide
Spectrophotometric Nucleic Acid Quantification in Microplates and Micro-Volumes
Nucleic acids are the building blocks of life in all living things, from plants and animals to bacteria and viruses. In research, it's important to quantify RNA and DNA prior to downstream processes like sequencing, restriction enzyme digestions and ligations, PCR and qPCR along with many other applications. Along with determining nucleic acid concentrations, it's also important to calculate the ratio of nucleic acid to protein to ascertain purity before using the sample in downstream applications.
There are fluorometric and spectrophotometric methods for nucleic acid quantification. The former is used for applications requiring high sensitivity due to minute available amounts of nucleic acid; the latter is more conventionally used from common nucleic acid extraction procedures. In this application guide, we'll discuss in detail spectrophotometric methodology as performed in microplate instrumentation and in specialized micro-volume accessories.
Product Spotlight
Epoch 2 Microplate Spectrophotometer
BioTek's Epoch 2 Microplate Spectrophotometer provides the ideal platform for nucleic acid quantification, whether you're working in microplates or cuvettes, or seeking to downsize your quantification assays to micro-volume samples. The monochromator-based optics enable wavelength selection from 200 – 999 nm in a little as 1 nm increments, so there's no need to purchase filters for any applications. Choose the protocol from the pre-defined options, or easily create your own through the touchscreen interface. Use the Take3 Micro-Volume Plate to miniaturize sample volumes to 2 µL, directly quantifying up to 48 samples at a time, without requiring additional instrumentation or software. Epoch 2 has excellent performance for nucleic acid quantification, protein quantification and many more research assays.
Tek Tips
Nucleic Acid Quantification in Microplates
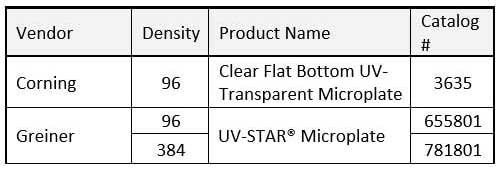
The simplest method for the quantification of nucleic acids uses the intrinsic chromophores provided by the purine and pyrimidine bases which possess a maximum absorptivity at 260 nm. Labware used for this measurement are varied and range from standard 1 cm path length cuvettes to micro-volume analysis where path lengths are typically 20x smaller. The common attribute to these labware is the need to be transparent to this UV light. Many plastics and glass will absorb 260 nm light, hence the widespread use of quartz for these measurements. Microplates are alternative labware, most useful when many samples must be analyzed. Due to their design, the optical path must be perpendicular and pass through the bottom of the wells. Rather than the use of quartz bottoms, microplate manufacturers incorporate UV transparent polymers to allow for the 260 nm measurement. As examples, the table below provides some commonly used microplate options for nucleic acid quantification.
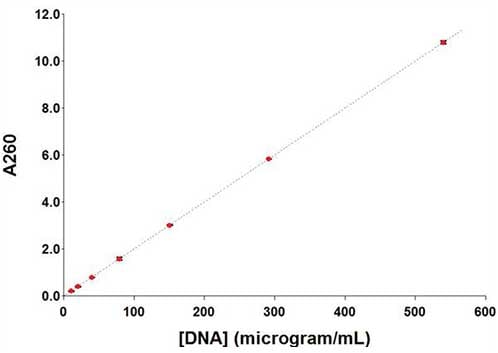
Another consideration in using microplates for nucleic acid quantification is that the path length through the sample will depend on the volume of the solution being measured. Given that most DNA samples will be aqueous in nature, path length correction to 1 cm can be made by measuring the absorbance of water at 977 nm (A977). BioTek's reader control and data analysis software, Gen5, has built-in methods for path length correction to 1 cm for samples in microplates where path length is defined by the volume of the sample added. The calculation is performed by measuring the absorbance peak of water at 977 nm and 900 nm, the difference is then divided by 0.18, which is the absorbance of water at 1 cm, equaling the sample's path length. In the figure below, BioTek's Synergy LX Multi-Mode Reader with absorbance monochromators was used to measure absorbance at 230, 260, 320, 900 and 977 nm in a single protocol to allow automated path length correction and accurate DNA quantification.
Nucleic Acid Purity Assessment
The assessment of the purity of a nucleic acid sample is often performed by a procedure commonly referred to as the A260/A280 ratio which refers to two spectrophotometric measurements made at these defined wavelengths. Pure nucleic acid samples typically have an A260/A280 ratio of 2.0. The absorbance of samples that contain a mixture of DNA and protein or other contaminants will be influenced by all molecules present and the ratio will be expected to deviate from the expected ratio. BioTek's range of hybrid microplate readers provide several read options including filter and monochromator based absorbance and fluorescence as well as luminescence measurements over a broad range of sample volumes and vessel types.
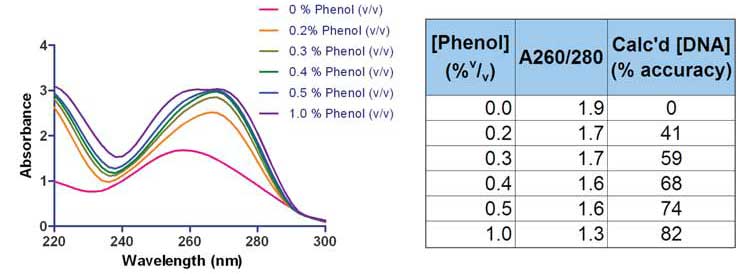
For example, spectral analysis for detection of the presence of residual phenol, a commonly used nucleic acid extraction reagent, shows obvious changes in the A260/A280 ratio as seen in the figure and table below. Thus providing a quick indication of potential contamination.
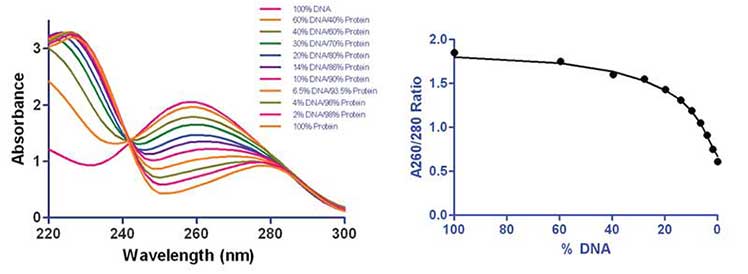
However, that may not always be the case. For example, in the spectra below, biomolecules were scanned to capture the absorption profiles of biomolecules as well as those of any potential contaminants, should they possess a chromophore of significant absorptivity over this wavelength range. When the A280 ratios were plotted against the varying concentrations of DNA/Protein (w/w) it is obvious that the A260/A280 ratio is reduced with increasing protein contamination. However, note that it takes a significant amount of protein contamination before a meaningful change is noted in the ratio (~40%).
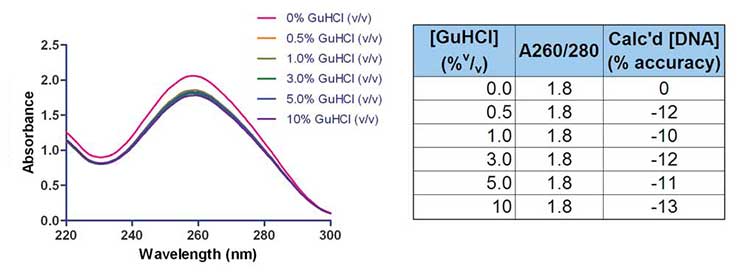
This is also the case for contamination with residual guanadine hydrochloride, a commonly used chaotropic agent, as seen in the figure and table below. While significantly effecting the accuracy of the quantification of DNA the contaminant is not recognizable by examining the ratio alone.
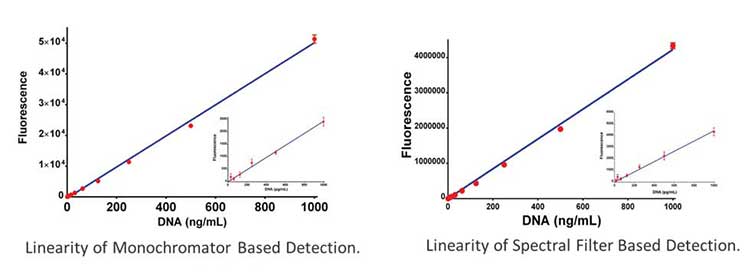
Therefore, it may be prudent to employ a target specific reagent for detection of more complex samples or for validation of upstream processes to insure removal of residual contaminants. For example, by using the dsDNA specific reagent Picogreen in conjunction with fluorescent filters or monochormators an accurate determination of dsDNA concentration can be obtained as seen in the figure below. These determinants than can be compared with absorbance based measurements and any discrepancies can be evaluated for potential causes.
Nucleic Acid Quantification Resources
Application Notes
- Fluorometric Quantitation of dsDNA using PicoGreen®(Cytation 1, Cytation 5, Synergy HTX, Synergy Neo2)
- Low-volume, High Throughput Workflow for Analysis of Nucleic Acid Samples for Biobanking (Synergy Neo2)
- Micro-Volume Purity Assessment of Nucleic Acids using A260/A280 Ratio and Spectral Scanning (Epoch 2, Epoch)
- Analytical Performance of a Higher Throughput Micro-Volume Microplate Accessory for Microplate Spectrophotometer Platforms (Epoch 2)
- Micro-Volume PicoGreen® DNA Quantification using BioTek's Take3 Multi-Volume Plate (Synergy HTX)
- Multi-Volume Analysis of Nucleic Acids Using the Epoch Spectrophotometer System (Epoch)
- Low Volume Nucleic Acid Quantification using a Multi-Volume Spectrophotometer System (Epoch)
Visit our Nucleic Acid Quantification application page to see more Nucleic Acid Quantification applications from BioTek instruments.
For Research Use Only. Not for use in diagnostic procedures.