Microbiology
July 2020
We are continually increasing our understanding of the significant roles microbes play in our lives, both as beneficial organisms and powerful research tools, as well as their involvement in numerous disease processes. Microbiology applications are numerous, including diagnosis and treatment of animal and plant disease, food and water safety testing, production of organic compounds, environmental health and impact appraisal, horticultural and agricultural soil assessment, food and beverage production, and exploration of alternative fuel sources.
Characterizing the highly diverse array of microorganisms requires reliable culturing techniques coupled with sensitive detection methods and analysis. The LogPhase 600 Microbiology Reader is designed for measuring microbial growth curves in up to four standard 96-well microplates at a time. LogPhase 600 features purpose-built, robust shaking and consistent temperature control, which are critical to optimal bacteria and yeast cell growth, for the best data quality. LogPhase 600 is controlled with a software App that provides a simple, yet powerful, user experience. The LogPhase 600 App controls the reader to acquire data and perform microbiology-focused analysis for all plates.
BioTek's Lionheart Automated Microscope and Cytation Cell Imaging Multi-Mode Readers support a broad range of microbiology applications. Fluorescence and luminescence measurements provide quantitation of microbial culture density and viability, while label-free and fluorescence imaging enables detailed classification and profiling of yeast and bacteria cultures using direct cell counts and subpopulation analysis.
Featured Applications
Monitoring Bacterial Growth under Different Environmental Conditions

Microbial organisms, such as bacteria, serve critical roles in many different facets of modern day living. They are used for the production of medical products, environmental remediation, biomedical research, and food production, to name just a few different uses. The ability to manipulate and select different strains with specific characteristics has been paramount to their utility. Likewise, the development of new strains often requires monitoring the growth of strains under various conditions. Here we describe the use of the LogPhase 600 Microbiology Reader to provide temperature control, suspension agitation, and monitor bacterial growth using light scatter in 96-well microplates.
Multi-Mode Metabolic Profiling of Stationary Phase Saccharomyces Cerevisiae Exposed to Short-Term Oxidative or Nutrient Stress

Due to the prevalent use of yeast as a model system, the choice of genomic, biochemical, and cell biology tools compatible with these cells is well-developed and wide-ranging. The ability to choose from a variety of assays that can combine optimal workflow productivity with relevant data outcomes can be limited by the need for multiple devices to perform different task and detection methods. Here we demonstrate a flexible and versatile instrument that supports a broad range of yeast studies, combining multiple imaging and detection modes with on-board environmental control and shaking. In this study, multiple assay formats were used to conduct a detailed characterization of oxidative and nutrient stress response in Saccharomyces cerevisiae.
Product Spotlight
LogPhase 600 Microbiology Reader
The LogPhase 600 Microbiology Reader captures and measures microbial growth curves in up to four 96-well microplates at a time. It features robust shaking and consistent temperature control for optimal bacteria and yeast cell growth. The LogPhase software app provides a simple, yet powerful, user experience.
BenchCel Microplate Handler
The BenchCel Microplate Handler is integrated to a variety of BioTek's microplate washers, dispensers, readers and imagers to automate application-specific workflows. The system is modular and scalable to suit a range of applications including automated ELISA, add/read protocols and cell fixing, staining and imaging processes.
Tek Tips
Tips for Automated Imaging of Yeast
Vessel Selection and Protocol Options
One of the most important considerations when imaging yeast and other microorganisms, especially when doing live cell imaging, is choosing the best vessel for your application. As these organisms are often in suspension your goal for imaging and analysis is to physically constrain the cells in the Z direction, particularly for kinetic imaging workflows. Although Gen5 can automatically image through your sample in Z, limiting the depth you need to cover will increase speed of your workflow and limit the amount of data that is acquired for your experiment.
For lower throughput imaging applications such as cell counting, live/dead assays, or phenotypic characterizations of yeast we recommend using vessels such as our reusable cell counting chamber (hemocytometer) and adapter, standard microscope slides, or channel slides as shown below. One great option to increase the throughput of these assays would be to culture or inoculate chamber slides that allow you to remove the media and chambers followed by coverslip mounting for microscopy. This could potentially allow for 32 samples to be imaged if using an 8-well chamber slide and our 4-slide adapter.
To show you how this might look in your own experiments we have compared two vessels for imaging yeast. The image on the left shows a well in which the yeast cells are in focus at multiple Z heights and are aggregating quite heavily, whereas the image on the right shows the same cells constrained in a channel slide. The ability to count the individual cells would be much less reliable in the vessel that does not control the cells in the Z direction, whereas Gen5 could accurately quantify cells in the channel slide.
Here is a nice example of a yeast kinetic growth assay performed in a constrained vessel, providing a monolayer of cells, which will make downstream counting and analysis much simpler for you to optimize in Gen5.
For higher throughput microplate imaging of yeast we suggest first diluting your cells to a countable range if cells were grown in the microplates, or inoculating your wells with an appropriate dilution for accurate cell counting. Remember, you can always add the dilution factor in your Gen5 plate layout and/or add a data reduction step to automatically calculate your concentration of cells regardless of what vessel you choose to utilize.
The second thing you will need to do is to either 1) add a delay step to your protocol to allow for your cells to settle, or 2) gently spin your cells to the bottom before putting them on your instrument. The first option is particularly helpful when running longer kinetic assays in which your plate is not on the instrument the entire time. For those assays we suggest setting up a discontinuous kinetic protocol in Gen5 so that your time points append in one experiment file. If your imaging is endpoint and you have access to a microplate centrifuge, a quick spin will be able to speed up the settling process.
Finally, we also suggest using the “slower carrier speed” option with cells that are non-adherent if you are using a Cytation Cell Imaging Multi-Mode Reader for your imaging experiment, which will prevent disruption of your cells from the bottom of the plate. You can select this option in both image manual mode and when designing your protocol as shown below.
Analysis Considerations
When doing analysis on yeast and other single-celled organisms, especially for object level analysis like cell counting and phenotypic characterization, it is always important to properly segment your cells. Clumping or aggregation can occur in your samples, but with Gen5 software you can accurately separate touching cells with only a few changes in your analysis pipeline. The figure below shows the default object masking parameters using the Texas Red fluorescent channel on the top and the optimized masking on the bottom. Always start by adjusting main masking parameters such as threshold and size, below we used a threshold value of 7000 RFU to remove some dim artifacts from staining, and adjusted the size to only pick up objects between 3-15um. The most important parameter to adjust when attempting to segment cells that are very close or touching is the rolling ball diameter for setting the mask. You can find this in Gen5 by clicking on “Advanced detection options...” and de-selecting “Auto” under the background flattening step as shown in the figure. For something very small, like yeast or other microorganisms, set this number aggressively low like 2-5um for example. You can see that those three simple changes in the Cellular Analysis window in Gen5 allowed us to accurately count the individual cells.
A great strategy for your staining workflows is to try to use fluorescence when available, as this is much easier to optimize. The line profile tool in Gen5 allows you to view the intensity of objects and their background, and as you can see from the images below, the fluorescent images have much more defined peaks and valleys. To use this tool, select the line profile tool and simply drag the mouse across the image making sure to go through some cells. There is often autofluorescence in common stains like the Crystal Violet shown in these representative images and we highly recommend using that to your advantage if ever possible. You can still take your color brightfield images, but the analysis will be much more user friendly by using the fluorescent channel.
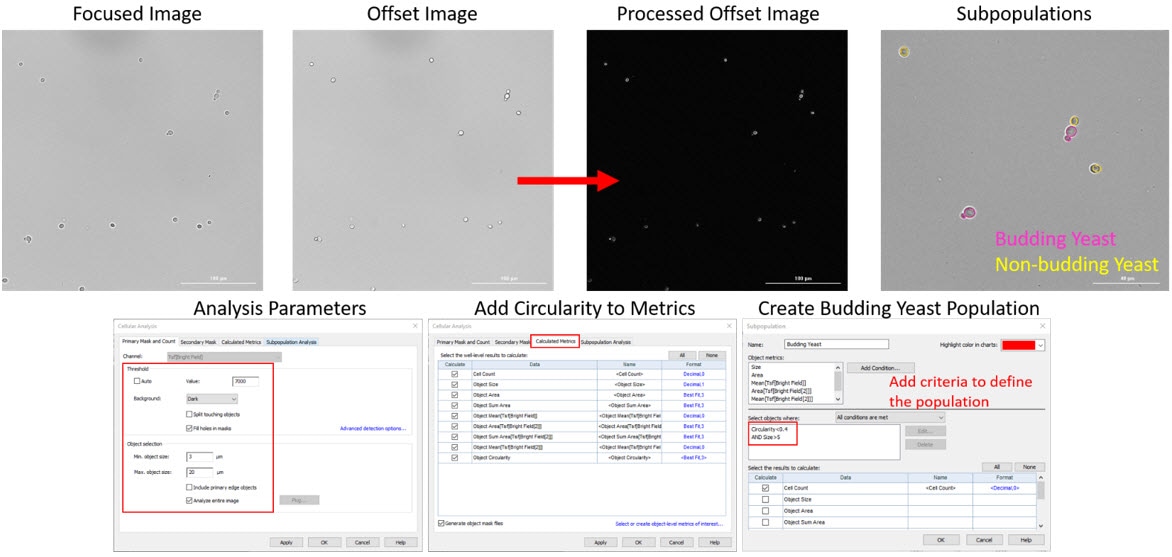
If you are interested in grouping your yeast cells into distinct groups, such as budding verses non-budding cells, you can use the subpopulation tool in Gen5 to create these populations. In this assay we have also utilized our high-contrast brightfield imaging kit to do this in a label-free workflow, which can be used with other microorganisms as well. You can simply take two brightfield images, one in focus and one slightly out of focus, the goal here being to illuminate the cells similar to a phase contrast image. After taking the two images processing is added automatically in Gen5 to “invert” the offset image to having a dark background with light cells, which will make it easier to define individual objects. We can add circularity as an additional calculated metric to help define the budding yeast population as they are not as circular as the non-budding population. For creating the subpopulation of budding yeast we used both circularity and size to distinguish these (in pink) from the non-budding population (in yellow) on the last picture below. This technique can be applied to create multiple subpopulations on a single image with varying parameters per group and if helpful when needed to filter out objects or debris that you do not want counted in your quantitative output.
Considerations for Optimizing LogPhase 600 Microbial Growth Studies
The LogPhase 600 Microbiology Reader performs kinetic OD600 determinations while providing temperature control and shaking for up to four separate 96-well microplates. When setting up experiments using the LogPhase 600 in addition to the experimental design, there are a few key elements that need to be addressed due to the vigorous shaking of the reader. First is the plate itself. Sterile flat bottom plates are highly recommended. It is incumbent for microbial growth studies that the only microbes being detected are the ones introduced as part of the inoculation process. The use of flat bottom plates allows for reliable OD determinations on the same well, read to read. While the LogPhase 600's location repeatability is quite good, there will always be some mechanical variability. If round bottom plates are used the physical path length of the material can vary depending on the location of the light beam.
We have also found that flat bottom plates provide more reliable mixing. We have found the Corning clear 96-well plate (P/N 3598) to be an excellent plate to use. These plates need to be covered with a true plate seal rather than the supplied lid. While the supplied lid works well for maintaining sterility, these plastic lids are quite loose and will wear with shaking. Over time the plastic dust generated can contaminate the inside of the reader. With short term or anaerobic experiments a standard optically clear impermeable seal works well. With longer experiments oxygen consumption can become an issue so a gas permeable seal can be used as long as they are optically clear. No need to worry about fogging, as the LogPhase 600 can employ a slight temperature gradient to eliminate condensation.
Lastly, the volume of the sample in the well is important. While a standard flat bottom 96-well microplate well can hold approximately 350 µL, it is important that the maximum volume used with microbial assays in the LogPhase 600 is 150 µL. Not surprisingly, the orbital shaking of the reader will cause the fluid to swirl in the well, with the fluid level sloshing upward around the well with the shaking orbit. With increasing microbial growth the surface tension of the fluid will decrease, as a result the degree of fluid sloshing increases dramatically. With too much fluid in the well, fluid will eventually splash on the plate seal, resulting in aberrant OD reading. We have found that by limiting the volume in the well to 150 µL, this phenomenon does not occur.
Microbiology Resources
Application Notes
- Automated Scoring of Acid-fast Bacteria (AFB) by Fluorescent Smear Microscopy
- Quantitative Image Analysis of Bacilli Viability in Agar Mounted Soil Samples
- Automated Image Analysis of Dictydium Sporothecae Biovolume Using Robotics
- Image Analysis of Herpesvirales Specificity Challenge using Direct Fluorescence Antibody Assay (DFA)
- Monitoring Viral Infection of Mammalian Cells using Digital Fluorescence Microscopy
- Monitoring Saccharomyces cerevisiea Growth with Brightfield Microscopy in Real Time
Tech Note
Webinar
Image Analysis 101 - Case Studies in Microbiology
Presenter: Wendy Goodrich, Applications Scientist
In this webinar, we'll describe how automated digital microscopy methods and high content image analysis tools frequently used in mammalian cell studies are applied to an array of microbial cell models common to bacteriology, virology, mycology, and mixed cell type co-cultures. Using live and fixed microorganisms, and a variety of stains, fluorescent probes and a label-free technique to demonstrate how downstream qualitative and quantitative data outcomes, such as cell counting and unit volume conversion, metabolic activity, strain differentiation, and morphology characterization, are obtained for each of a group of individual case studies. Our goal is to provide tips and insights that can be broadly applied in microbial imaging applications beyond the examples shown.
For Research Use Only. Not for use in diagnostic procedures.