Genotoxicity
January 2020
Quantifying DNA damage and accurately measuring the rate and extent of repair is an important aspect of a range of research fields, including epidemiology, toxicology, and drug development. Mammalian cells incur DNA damage as a result of internal factors, for example stress, oxidation and metabolism, as well as exposure to external stimuli, such as chemical compounds, UV and cosmic radiation. Genotoxicity studies aim to identify and understand the mechanisms of DNA damage and repair, and to better understand regulatory pathways to improve therapeutic success.
The comet assay is well established for quantifying DNA damage in mammalian cells and is compatible with both detection of a wide array of DNA damaging agents and monitoring the cellular DNA repair capacity for several different DNA repair pathways. The comet assay consists of DNA migration in an electrophoresis gel, where intact DNA (comet head) moves at a slower rate than fragmented DNA (comet tail) as evaluated via fluorescence staining and microscopy. The percentage of fragmented DNA in the comet tail is a direct measure of DNA damage.
Comet assays are typically performed and analyzed manually, so although the assay provides increased sensitivity, adoption as a reliable method has been slow. However, recent advances in automated imaging and analysis techniques have enabled high-throughput comet-based genotoxicity studies. Using this automated method, high-density slides allow 96 individual samples to be compared simultaneously. The combination of a novel single-cell microarray comet assay format, and automated instrumentation and analysis, provides an accurate, robust method to assess DNA damage and DNA repair in mammalian cells.
In mammalian cells, one of the most serious types of DNA damage is a double-stranded break (DSB), where the DNA double helix is completely severed. One of the most characterized early markers of DNA DSBs is the phosphorylation of the histone 2AX (H2AX) to γH2AX. Here, a positive feedback loop is created between γH2AX and phosphatidyl inositol 3' kinase-related kinases, where repair systems are recruited to the damage, creating nucleation sites, or foci, that correspond to the level of DSB. When the foci are bound to fluorescently labeled antibodies, overall γH2AX levels can be visualized and quantified via immunofluorescence microscopy.
The conventional, manual method of evaluating foci via a γH2AX assay offers limited throughput and is subject to operator variability. Incorporating automated imaging and analysis into the assay workflow can help to enhance throughput, assay robustness and accuracy, while also increasing overall laboratory efficiency.
Featured Applications
Automated Imaging and Dual-Mask Spot Counting of γH2AX Foci to Determine DNA Damage on an Individual Cell Basis

Double strand DNA breaks represent a critical form of genotoxic effect. Associated with cell death and tumorigenesis, they are well defined by the phosphorylation of histone 2AX (H2AX) to γH2AX as part of the DNA repair process. In the procedure described here, following immunostaining, automated fluorescent imaging and dual mask spot counting is performed to quantify labeled foci per nuclei after drug treatment.
Automated Comet Assay Imaging and Dual-Mask Analysis to Determine DNA Damage on an Individual Comet Basis

Mammalian cells incur DNA damage and alterations as a result of internal factors, such as stress, environment, oxidation and metabolism, as well as exposure to external stimuli, such as chemical compounds, UV and cosmic radiation. Genotoxicity studies aim to identify and understand the mechanisms of DNA damage and repair, and also to seek methods of pathway regulation and dysregulation for improved therapeutic success.
Product Spotlight
New Cytation 7 Cell Imaging Multi-Mode Reader
The latest innovation in our Cytation line of cell imaging multi-mode readers, Cytation 7 brings upright microscopy and inverted microscopy together with multi-mode detection. It is a powerful, patented combination that supports a very broad range of applications, from ELISpot imaging, slide scanning, 3D cell imaging and analysis, to live cell imaging and biochemical assays. Visit our product page to learn more about why Cytation 7 is truly Ready for Any Assay!
Watch this video to learn more about the complete Cytation line of cell imaging multi-mode readers.

Tek Tips
Optimizing image processing steps and dual masking settings
Image analysis of genotoxicity assays, such as comet and γH2AX immunofluorescence assays, often requires advanced quantification via dual masking. Dual masking allows identification of objects in two channels associated with each other. For example, in comet assays the primary mask is placed on the comet head in each image and a linked secondary mask is placed around the entire comet to identify the tail. This allows for quantification of both undamaged (head) and damaged (tail) DNA.
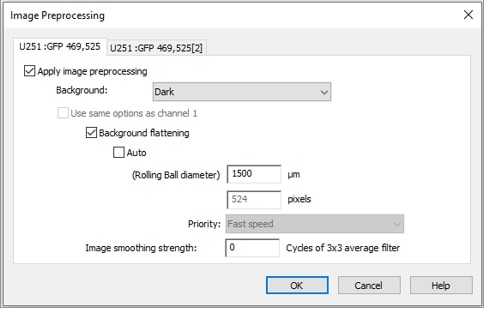
When applying image preprocessing prior to analysis, choose a rolling ball diameter large enough to ensure that enough of the image area is sampled for distinguishing background from comet signal. Since some comet tails can have weak signal, using a rolling ball diameter that is too small can cause removal of true signal from the comet tails, thereby giving false results.
Primary masks should then be applied to the first channel to identify comet heads, followed by secondary masking on the second channel of entire comets for tail analysis. These two “channels” may be the same fluorophore (for example, both GFP channels), but you must capture two overlaid images in the procedure in order to apply secondary mask analysis.
The use of dual masking for analysis allows each comet head and tail to be properly linked, improving accuracy of results. If overlapping comets, debris, or other objects are present in the image that produce false results, subpopulation analysis may be used to exclude these artifacts. Common calculations of desired data output, such as percent DNA in the tail and comet tail moment, can be added to a Gen5 protocol for automatic reporting. Properly optimized analysis parameters yield consistent, unbiased, accurate data. These optimized settings then can be easily and repeatedly applied to comet assays in an automated, high throughput format. The final results reveal the extent of DNA damage from an imaged sample, where comet heads appearing as well-defined spheres indicate little damage while comets with distinct tails indicate extensive damage, allowing both analysis of individual comets as well as population level results.
Gen5 spot counting analysis for accurate evaluation of γH2AX assays
Quantifying small foci, or spots, of fluorescence is required when performing γH2AX assays. Spot counting of foci associated with individual nuclei adds another layer to secondary mask analysis (note: Gen5 Image Prime Spot Counting add-on module required). Primary masks are first added to automatically identify nuclei in each image, while secondary masks identify spots of DNA damage. Aggressive preprocessing of the spot counting channel prior to masking is used to remove as much background fluorescence as possible, leaving behind bright foci of DNA damage. This allows for accurate analysis of individual labeled spots in each nuclei.
When adding an image preprocessing step, keep your primary channel as usual for analysis (either default settings with auto rolling ball diameter or with your usual custom settings for identifying cells), but change the rolling ball diameter for the secondary, spot counting channel. Select your secondary channel tab, uncheck “use same options as channel 1”, uncheck “auto”, decrease the size of the rolling ball diameter (usually 1-2 µm for a single image), and change priority to “fine results”.
Gen5 offers several options to identify areas of interest for secondary mask spot counting, including within the primary mask, outside of the primary mask, and reducing or expanding the primary mask. Choose the option that best suits your desired area of analysis for spot counting, dependent on your assay. You can also adjust minimum and maximum spot size, threshold, and rolling ball size (available under “advance options”). Since even small modifications to the analysis parameters can make a big difference in spot counting, experiment with different settings to determine the best masking for your samples.
The high quality images captured on BioTek's Cytation and Lionheart automated imagers together with the powerful analysis capabilities of Gen5 position researchers for success in obtaining accurate, reproducable data from complex assays. As described above, the use of automated imaging and analysis via Gen5 Image Prime with dual masking and spot counting module creates a user-friendly experience with accurate results for genotoxicity experiments, including comet and γH2AX assays.
Genotoxicity Resources
Webinars
Automated Imaging and Analysis of Genotoxicity using Single Cell Gel/Comet Assay
Presenters: Sachin Katyal, PhD, Senior Scientist, Manitoba Institute of Cell Biology, CancerCare Manitoba; Brad Larson, Principal Scientist, BioTek Instruments, Inc.
In this webinar we demonstrate imaging and analysis procedures using the CometAssay® from Trevigen®. Following staining, automated imaging of each well is performed using the Cytation cell imaging multi-mode reader. Built in Gen5 software, used for analysis and instrument control, then carries out commonly accepted calculations; such as % DNA in the comet tail and tail moment. The combined process can then be used to generate automated, high throughput comet-based genotoxicity data.
For Research Use Only. Not for use in diagnostic procedures.