Cell Migration
June 2019
Cell migration is the movement of cells from one location to another, driven by complex environmental signaling events and induced structural changes to the cytoskeleton. Multicellular organisms rely on cell migration for a vast array of biological processes, including embryonic development, immune responses, and wound healing. Cell migration is also critically important to various pathological processes, notably cancer metastasis.
In this edition of TekTalk, we will focus on useful tools for conducting cell migration assays including a fully automated cell migration assay using the AutoScratch Wound Making Tool, which provides a convenient and reproducible method for quantifying 2D cellular migration in either 24- or 96-well formats; and a barrier assay using biocompatible gels for high throughput operation in 384-well formats. Additionally, we will present tips and tricks for optimizing cell migration assays.
Featured Applications
Advanced Wound Healing Assay Workflow using Automated Scratch Wound Creation, High Contrast Brightfield and Fluorescence Kinetic Imaging
Multiple components, including extracellular matrix and stromal cells, play a role in constructing in vivo tissues or cancer cell models. When performing scratch wound healing assays to determine the migratory patterns of target cell types included in these models, it is important to create the most in vivo-like setting for pre-clinical in vitro tests. The current standard for performing scratch wound assays involves manual wounding of a cell layer created on the bottom of a non-coated plate, and label-free cellular analysis. With the addition of an automated wound creation method, in addition to kinetic image capture and analysis of both brightfield and fluorescent signals from single and co-cultured cells, robust data can be generated to allow accurate assessments of potential promotion of wound closure or prevention of metastatic cell migration.
Automated Label-Free Method for Measuring Cell Migration in Real-Time with Oris Pro Assay
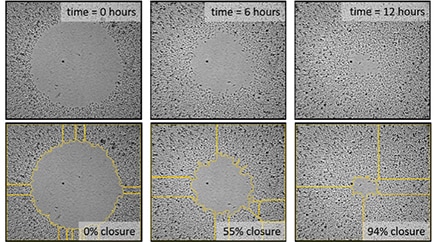
Cell migration is dependent on complex environmental signaling events and induced structural changes to the cytoskeleton. Multicellular organisms rely on cell migration for a vast array of biological processes, including embryonic development, immune responses, wound healing, and cancer metastasis. Here we demonstrate an automated kinetic imaging-based approach to investigating cell migration. This convenient label-free method enables accurate and robust analysis of cell migration in 96- or 384-well microplates for high-throughput applications.
Product Spotlight
Cytation 5: Now with Wide Field of View Camera
Cytation 5 Cell Imaging Multi-Mode Reader is now available with a wide Field of View (wide FOV) camera. The wide FOV camera enables a single image capture of an entire well of a 384-plate, and also facilitates rapid image capture for large samples. It is ideal for capturing rare event biology, as seen in assays such as circulating tumor cell assays, plus more statistically significant sampling required for assays such as transcription factor activation. Along with Gen5 software for image processing and analysis, the new camera for Cytation 5 provides even more functionality for broad, live cell imaging research.
Scratch Assay Starter Kit
Designed for simple, automated and repeatable implementation of cell migration assays, BioTek's new Scratch Assay Starter Kit provides everything a researcher needs to implement automated, kinetic cell migration and scratch wound assay. Used in conjunction with BioTek's Lionheart or Cytation imager, the Starter Kit includes: BioTek's AutoScratch Wound Making Tool to create repeatable scratch wounds, the Scratch App software with pre-defined protocols to automatically calculate key statistics including wound width, % confluence and maximum healing rate, sample packages of 24-well and 96-well microplates, plus reagents to ensure the best scratch performance and best scratch assay analysis.
Tek Tips
Automated cell migration analysis
Migration is a key property of living cells and critical for normal development, immune response, and disease processes such as cancer metastasis and inflammation. In vitro cell migration assays are very useful and important for a wide range of biomedical research such as cancer biology, vascular biology, cell biology and developmental biology. The in vitro wound healing and transwell migration and invasion assays are widely used in the scientific community to assess the cell migration.
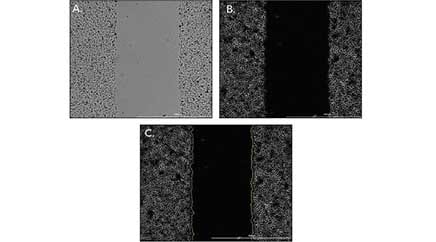
BioTek's solution for automated wound healing assays Among the various methods to study cell migration, scratch/wound healing assays are widely used in the scientific community. This method is useful to determine the migration ability of the whole cell mass. Measuring the wound closure over time when compared to a control may reveal specific migration changes or an impaired migratory phenotype. Currently, wound healing assays are mostly performed manually. With this method, cells are seeded to reach a confluence of 90-100%. Pipette tips are then used to manually create a scratch in the monolayer. In spite of the perceived simplicity of these manual techniques, created wounds are subject to high variability imposed by users. For instance, depending on the force applied by the user, the width of the gap can vary across the replicates. In addition, excessive force against the tissue culture plate with the pipette tip can damage the coating of the surface or the cell monolayer itself. With the addition of an automated wound creation method, consistent and reproducible scratch wounds can be created across the wells of multi-well plates to generate robust data for accurate assessment of the potential impact of target compounds on migration. In addition, automated kinetic imaging and quantitative analysis of captured images using BioTek's automated imaging platforms (Lionheart FX, Cytation 5, and Cytation 1), enhances the speed of performing the experiment and minimizes the user error in the final data. BioTek's AutoScratch tool can be used to automatically create reproducible wounds in both 96- and 24-well plates. A built-in washing routine keeps the scratch pins free of cell build-up and reduces both the chance of contamination, especially for co-culture experiments and the error in wound area measurements caused by cell build-up in the gap region. The latter concern can be further alleviated by adding a post-wound washing step using MultiFlo FX to remove floating cells that could confound analysis. To analyze overall migration of cells, images can be captured in either high contrast brightfield or normal brightfield channels (depending on which is available in your imaging platform). Captured images should be pre-processed to enhance the contrast between the background and cell occupied areas. For best results, it is recommended to use a small rolling ball diameter (20 µm) in Gen 5 software. However, this number can change when cells are seeded at a lower density or hasn't reach the desired confluence (~90-100%).
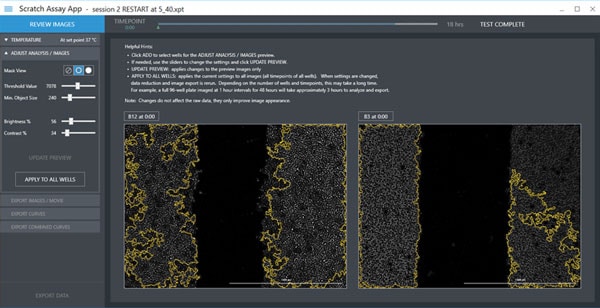
Cellular analysis can be automatically applied to the processed images to calculate wound area. Using the threshold slider, a correct threshold can be selected to separate two regions: the low-intensity open area and the high-intensity cell covered area. Gen5 allows the user to type the correct intensity threshold that results in the best segmentation. For AutoScratch users, four distinct sliders are offered in the Scratch Assay App: threshold value, minimum object size, brightness and contrast. The optimal image analysis settings required for computing the open area can be simply found by adjusting the sliders for those four different parameters. Before applying the setting to all replicates, it is advised to check the wound area selection for multiple wells especially if they belong to different samples. If the designated settings failed to identify the open area, users can further modify the setting such that a universal setting is found that works for all conditions. Gen5 software and AutoScratch give an option to manually mask certain wells in the experiment if the final threshold setting fails to accurately segment the wound area in those wells.
Minimizing proliferation To minimize cell proliferation, it is recommended to serum starve the cells for at least 24 hours before generating the scratch wound. Minimum concentration of serum at which cells can survive is completely cell-line dependent, and has to be determined experimentally. Alternatively, DNA synthesis inhibitors such as Mitomycin C can be used to inhibit the proliferation during the wound healing assay. Both of these methods will inhibit the cells' ability to proliferate.
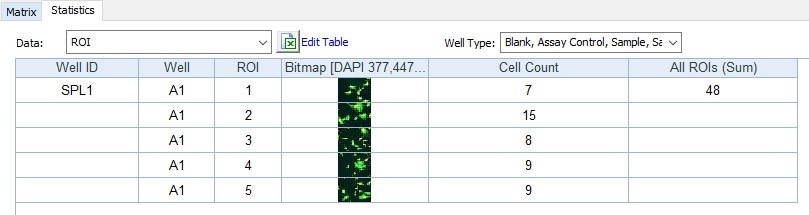
Automated kinetic imaging of cell migration through Transwell permeable supports The Transwell migration and invasion assays may be used to analyze the ability of single cells to directionally respond to various chemo-attractants. In this assay, cells are plated on the permeable support by adding a cell suspension to the apical compartment of the transwell insert. Reagents can be added to the basolateral compartment to induce chemotactic behavior in different cell types. The particular pore size of the permeable support used is dependent on the individual size of the cells in the sample. For example, if the cells are too small they may pass through the pores without the need to facilitate migration; or if they are too large they may not fit through the pores. There are several Transwell inserts with pore sizes that range from 3 µm, 5 µm, and 8 µm for cell migration assays. Correct cell seeding density and homogenous cell distribution across the permeable support is crucial for the success of the Transwell migration assay. Overseeding can cause colonization and will adversely affect cell migration through the membrane. However at lower density, cells don't form cell-cell junctions and the observed behavior will be more similar to single cells and not cell clusters. FluoroBlok cell culture inserts are designed with a polyethylene terephthalate (PET) membrane that efficiently blocks the transmission of light between 490 and 700 nm, allowing fluorescence detection in a simplified and non-destructive manner. Fluorescently labeled cells present in the apical compartment of the insert are shielded from bottom-reading microplate readers or microscopes enabling non-destructive kinetic and endpoint migration and invasion assays. Fluoroblok inserts are available in a wide array of formats and configurations from individual inserts to 96-well plate. Regardless of the positioning of the insert in the wells, a reading step can be defined to automatically monitor the migration through the Transwell permeable support. For inserts that are not placed in the center of the well, a beacon can be defined in Gen5 to automatically locate the insert inside the well. Montaged images can be taken to capture the entire membrane.
Imaging Regions of Interest in Gen5
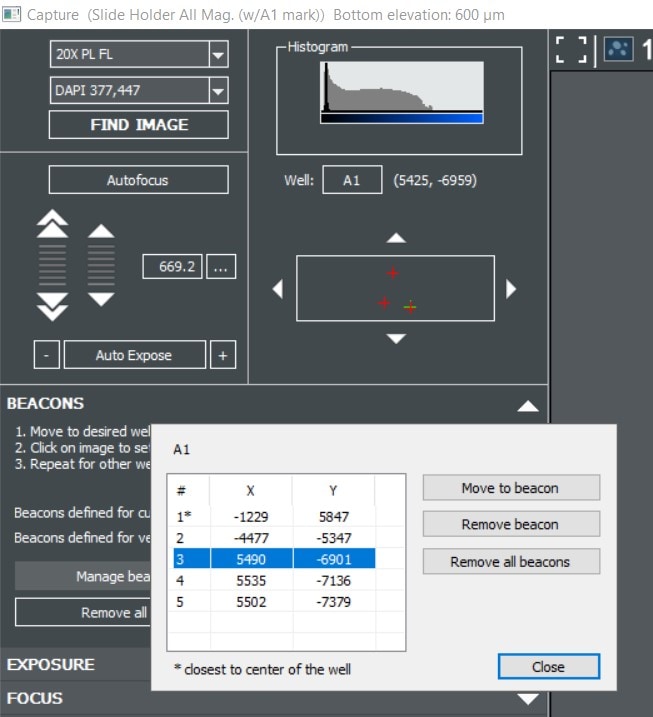
Gen5 offers several tools to automate high resolution imaging in microplates, slides, petri dishes and various other formats. These tools allow you to follow many samples in parallel over a kinetic time course while generating movies and monitoring biological changes. But what about a sample that is not uniformly distributed in a well? Or a rare cell type that makes up only a tiny fraction of your population? For example, an anesthetized zebrafish or a mitotic cell preparing to divide rarely lines up with the exact center of a well. For these situations, Gen5 allows you to manually define regions of interest on a well by well basis using imaging beacons.
New in Gen5 v3.06 you can define multiple beacons per well to make sure you capture the perfect field of view for each of your samples. When setting up your imaging step, check the “Define Beacons” option and then navigate through your sample to define and store up to 255 regions of interest. Images for each beacon are captured at your desired interval and can be analyzed individually or their results pooled to increase replicate numbers.
Cell Migration Resources
Application Notes
- Incorporation of a Novel, Automated Scratch Tool and Kinetic Label-Free Imaging to Perform Wound Healing Assays
- Automated Monitoring of Protein Expression and Metastatic Cell Migration using 3D Bioprinted Colorectal Cancer Cells
- Automated, Kinetic Imaging of Cell Migration and Invasion Assays using Corning FluoroBlok Permeable Supports
- Imaging and Analysis of a High Density Cell Migration Assay
- Performance of a Label-Free Image-Based 2D Scratch Wound Healing Assay to monitor Cell Migration and its Inhibition
Visit our Cell Migration application page to see more Cell Migration applications from BioTek instruments.
Webinar
A fully automated solution for conducting cell migration assays using the AutoScratch Wound Making Tool
Presenter: Joe Clayton, PhD., Principal Scientist

Cell migration is the movement of cells from one location to another, driven by complex environmental signaling events and induced structural changes to the cytoskeleton. Multicellular organisms rely on cell migration for a vast array of biological processes, including embryonic development, immune responses, and wound healing. Cell migration is also critically important to various pathological processes, notably cancer metastasis.
In this webinar we will demonstrate a fully automated cell migration assay using the AutoScratch Wound Making Tool (part of the Scratch Assay Starter Kit), which provides a convenient and reproducible method for quantifying 2D cellular migration in either 24- or 96-well formats. Additionally, we will present tips and tricks for optimizing scratch assays, including sample preparation, post scratch washing procedures, and techniques for ensuring accurate and meaningful quantitative results.
For Research Use Only. Not for use in diagnostic procedures.