Cell Counting
December 2018
Cell counting is a basic laboratory need for any research using cultured cells. While confluence measurements are sufficient to judge when to split cell cultures, an accurate assessment of cell numbers is required for cell-based assay development. Traditionally, these measurements are performed using a hemocytometer and brightfield microscopy where individual cells are counted on a grid and viability assessed through the use of a stain like trypan blue. This is a laborious, but inexpensive process. Digital microscopy and the use of image analysis algorithms to automatically count cells can simplify the process dramatically. Furthermore, similar algorithms can be devised for a wide range of quantitative microscopy applications where cells can be counted based on the expression of a desired phenotype. Often this employs a fluorescent probe that illuminates the phenotype of interest.
In this edition of TekTalk, we describe the use of cell counting for a range of applications from label-free cell counting for cell proliferation, nuclear stain-based cell counting for the normalization of cell-based data such as oxygen consumption and extracellular acidification rates and imaging assays monitoring the progression of apoptosis and necrosis through the use of selective fluorescent probes.
Featured Applications
Kinetic Proliferation Assay Using Label-Free Cell Counting
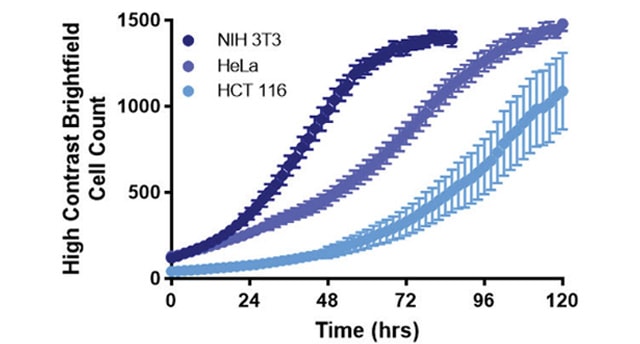
Kinetic cell proliferation assays enable quantitative analysis of cell viability and growth rates and are a powerful tool for evaluating cellular response to drug treatment. Conventional methods rely on either indirect biochemical measurements or the use of fluorescent labels. Alternatively, high contrast brightfield cell counting provides a direct and label-free method for measuring cell population size. Combining the high contrast brightfield imaging capabilities of the Cytation 5 Cell Imaging Multi-Mode Reader with the BioSpa 8 Automated Incubator enables convenient and robust long-term kinetic cell proliferation studies.
Live Cell Imaging of Apoptosis and Necrosis
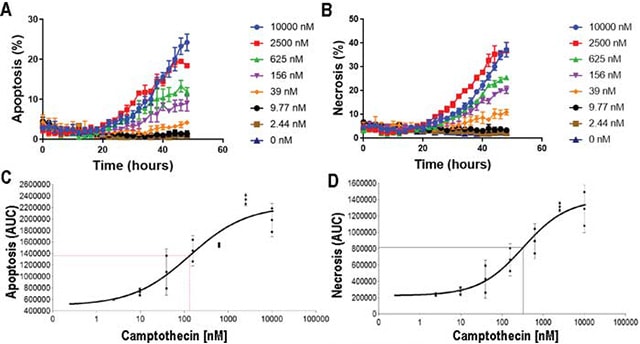
Cell death occurs throughout the life of an organism, and this is critical for developmental plasticity and organismal health, in part by eliminating unneeded and unhealthy cells in a timely and effective manner. However, dysfunctional cell death leads to diseases such as cancer, neurodegeneration, and ischemic damage. In this study, we investigate apoptosis and necrosis in real time over a 48 hour time period using high contrast brightfield imaging in combination with fluorescent probes for apoptotic and necrotic cell death markers. We demonstrate apoptosis and necrosis are induced in a dose dependent manner in both HT1080 and SKOV3 cells.
Partner Spotlight
Agilent and BioTek Collaborate on Seahorse XF Normalization
The power of the well-known Seahorse XF technology from Agilent Technologies lies in its ability to simultaneously and kinetically measure mitochondrial respiration and glycolysis - two major energy pathways - in live cells. By doing so, researchers can closely monitor these dynamic events in real time rather than using separate assays for each cellular activity. As mitochondrial respiratory rates correlate to cell number, small differences in the number of adherent cells present in each microplate well create differences in the data gleaned from the Seahorse XF workflow. Normalizing XF data allows researchers to compare experimental data from well-to-well, treatment-to-treatment, plate-to-plate and more.
Product Spotlight
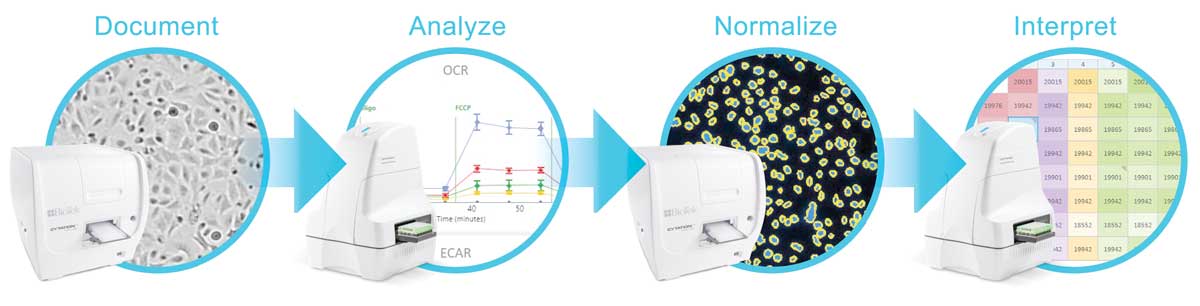
Cytation 1 and 5 Imagers - Automated Normalization Through Cell Counting
Seahorse XFe24 & XFe96 Analyzers are preeminent instruments for the kinetic measurement of cellular respiration and glycolysis in live cells. These measurements correlate to the cell number per well similar to other bioassays, such that normalization of results to cell number provides improved accuracy and precision to the data. In addition to the cell number, errors in sample preparation such as uneven seeding or partial cell loss during washing steps commonly result in outlier wells, which often cannot be corrected by data normalization.
Agilent and BioTek Instruments have developed an integrated platform that provides seamless normalization of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) data using BioTek's Cytation 1 and 5 Imagers to perform fluorescence imaging-based cell counting. Furthermore, brightfield imaging with the Cytation before Seahorse analysis discovers any sample preparation errors allowing the justified exclusion of outlier data. Both brightfield and fluorescence imaging is performed within Agilent's Seahorse XF WAVE software providing a seamless workflow:
- Brightfield imaging by Cytation 1/5 Documents wells providing a means to discover any sample preparation errors
- Cells are Analyzed by the Seahorse XFe Analyzer providing kinetic OCR, ECAR measurements
- Fluorescence imaging by Cytation 1/5 Normalizes the OCR, ECAR measurements to cell number
- Seahorse XF WAVE software Interprets the results providing accurate and precise measurements of cellular respiration and glycolysisstructures.
Tek Tips
Generating Accurate Counts of Immune Cells
Immune cells pose unique challenges for image-based cell counting applications that can be addressed with a combination of cell culture techniques and optimized image analysis settings. For example, a quick, low speed spin of suspension cells immediately after seeding in a microplate promotes attachment to the culture surface – allowing the cells to be captured in a single image plane. For populations consisting of cells ranging widely in size and intensity of nuclear staining, such as PBMCs, use the Gen5 cellular analysis tools to decrease the minimum object size and set the threshold to a fixed value low enough to include all cells. Additionally, for cells that are tightly packed together, consider more aggressive background flattening (e.g. rolling ball diameter of 3 mm) to separate and count individual cells within clusters (figure 1). Finally, images acquired with a 4x objective capture a large field of view while delivering the resolution and sensitivity necessary to count cells under most conditions. However, for more challenging cell counting applications, the increased resolving power and sensitivity of the 10x objective may be preferable.
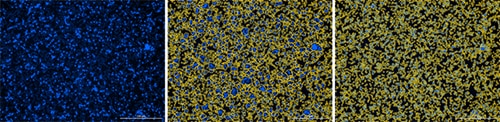
Figure 1. Accurate counts of immune cells can be generated using Gen5 image analysis tools. (A) PBMCs stained with Hoechst 33342 were seeded in a 96-well plate at a high density and spun down to attach cells to the well bottom prior to imaging with a 4x objective. (B) Tightly packed clusters of cells can be identified as single objects, resulting in artificially low cell counts. (C) Treating the image with aggressive background subtraction enables separation of individual cells and accurate counting.
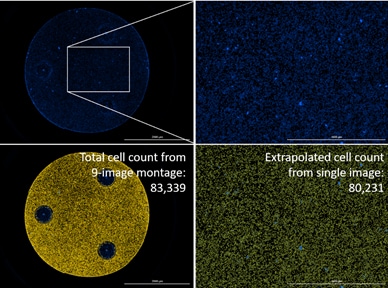
Efficiently determine total cell counts using a single image
Frequently researchers want to determine the total number of cells within a culture dish or microplate well for a wide range of applications, including data normalization across samples and time points. Often this does not require imaging the entire culture area as long as cells are relatively evenly dispersed. Gen5 cellular analysis and data reduction tools can be used to automatically calculate and report total cell counts within a well from a single representative image (figure 2). Optimizing cell seeding techniques, including leaving seeded microplates at room temperature for approximately 15 minutes prior to placing them in an incubator, will result in more evenly distributed cells and enable more efficient cell counting.
Real-time Cell Confluence Measurements
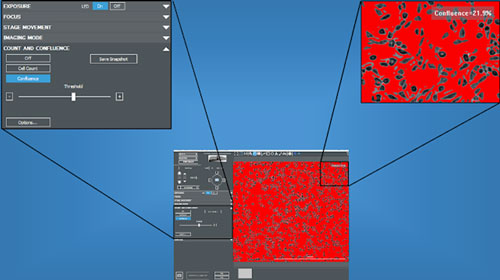
Sometimes in the lab you just need to get a quick record of cell count or confluence to determine if and when to move onto the next step in your workflow. BioTek's Gen5 software in combination with Lionheart Automated Cell Imagers or Cytation Cell Imaging Multi-Mode Readers provides a number of real-time measurements to be made rapidly in real time. Measurements can be made using any available objective and channel including high-contrast brightfield, phase contrast and fluorescence. In the image below the "Find Image" feature in Gen5, which includes Autofocus and Auto Expose features, has been used resulting in dark cells on a light background (Figure 1). This can then be used to determine cell confluence. It is critical to insure the correct plate type and bottom elevation within the plate imaging parameters are appropriately selected prior to performing image acquisition.
By selecting "Count" and "Confluence" from the available menu items in the Manual Mode control panel and selecting "Confluence" the calculation is performed instantaneously and the desired threshold can be adjusted in real-time to insure all objects are appropriately measured and background excluded (Figure 2). The result below indicates excluded background highlighted in red and a cell confluence of 21.9% for a sample in a 96-well plate format.
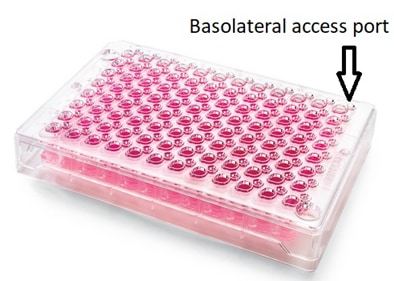
Washing Permeable Support microplates with off-center access ports
Permeable Support microplates are used for testing drug absorption/transport. These microplates consist of an insert plate containing a porous membrane that fits into a receiver plate such that two distinct compartments are formed in each well. Cells are then seeded into the wells allowing formation of a cell membrane which can be used as an in-vitro model for how orally taken drugs may be absorbed in the gut. The compartment above the cell monolayer represents the lumen of the gut and is thus named the apical compartment; that below the cell membrane represents the bloodstream and is thus named the basolateral compartment. The apical compartment can be accessed through normal means since it is openly accessible to standard liquid handling devices. The basolateral access is done through an access port on the insert plate. BioTek had developed Application Notes for working with these types of plates – specifically the HTS Transwell from Corning Life Sciences.
BioTek's washers and dispensers have been validated to work with the HTS Transwell plates and can easily access both compartments with some optimization of the wash/dispense protocols. The key parameter is setting up the offsets for the wash and dispense steps.
A recent customer had used another vendor for their Permeable Support microplates that have a different format than the Corning Transwell microplates.
Unfortunately, the issue with this microplate format is that the basolateral access port is outside of the positioning limits for a standard 96-well plate.
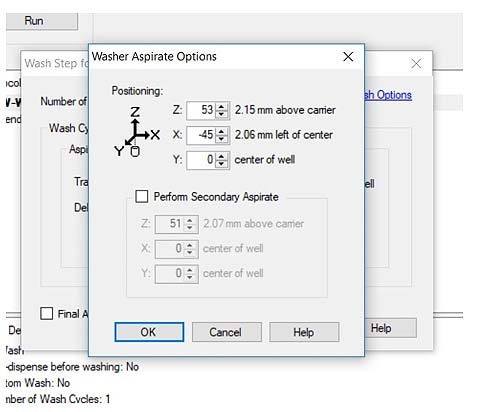
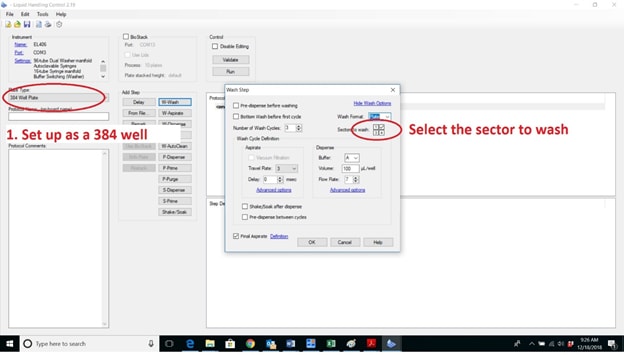
BioTek's washers and dispensers can work with these microplates if the wash setup is changed to 384-well microplate – this setting extends the limits of the pin positions and allows access to the port of the 96-well plate. The following example is shown in Liquid Handling Control (LHC) software. Disable the other sectors of the microplate so the wash protocols only access the appropriate one. You will still need to optimize the dispense and aspirate parameters to center the manifold over the wells.
Cell Counting Resources
Application Notes
- Live Cell Imaging of Apoptosis and Necrosis (BioSpa 8, Cytation 5)
- Automated Cell Counting with High Contrast Brightfield and EVE Counting Slides (Cytation 1, Cytation 5, Gen5, Lionheart FX)
- High Contrast Brightfield (BioSpa 8, Cytation 5, Gen5)
- Automated Hemocytometer-Based Live/Dead Cell Counting using Phase Contrast and Color Brightfield Imaging (Cytation 5)
- Analysis of Nuclear Stained Cells - Using the Cytation3 Cell Imaging Multi-Mode Microplate Reader with DAPI-Stained Cells (Cytation 1, Cytation 5)
Visit our Cell Counting application page to see more Cell Counting applications from BioTek instruments.
For Research Use Only. Not for use in diagnostic procedures.