About SureFISH Probes
_1.0.jpg)
|
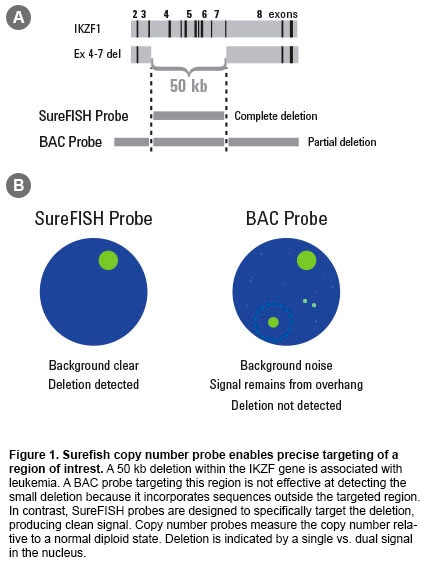
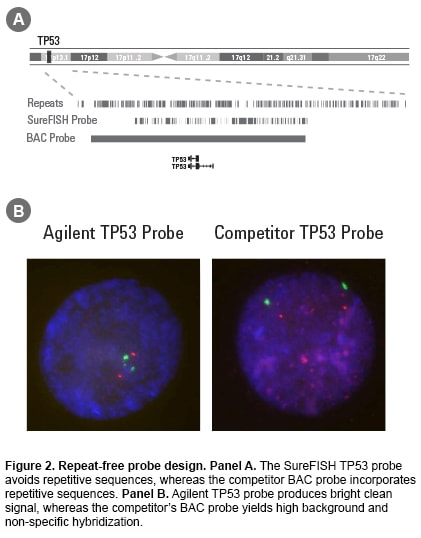
Fluorescence in situ hybridization (FISH) is a laboratory procedure in which fluorescently labeled DNA fragments (the FISH probe) hybridize to complementary DNA sequences in the cell’s nucleus. The resulting fluorescent signals are visualized using a microscope and indicate the presence and localization of one or more targeted DNA sequences. Agilent offers three types of probes: break apart, dual fusion, and copy number. Oligoucleotide–based FISH Probes Agilent’s unique SureFISH DNA FISH probes are designed in silico and chemically synthesized using the company’s high-fidelity, oligonucleotide library synthesis (OLS) technology. This eliminates the limitations of FISH probes manufactured with bacterial artificial chromosome (BAC) technology. Agilent is the only DNA FISH probe supplier that does not use BAC technology in probe manufacturing. High Probe Specificity—Precisely Target Designing probes on the computer “in silico” means complete design flexibility. An oligonucleotide (oligo) probe can be designed to precisely target any region of interest with base-pair level resolution (Figure 1). Precision probe design means higher probe specificity. High Signal-to-Noise Ratio—Free of Repetitive DNA SureFISH probes, because they are comprised of chemically synthesized oligos, do not suffer from high hybridization background and non-specific signal. To ensure that every SureFISH probe is truly repeat-free and will produce clean signal, each oligo is sequence compared to the entire human genome to verify target specificity. All non-specific sequences are removed. Thus, SureFISH probes eliminate the need for blocking and the signal suppression produced by blocking agents (Figure 2). High Lot-to-lot Consistency—Free of Cell Culture Agilent’s synthetic SureFISH probe production process ensures high lot-to-lot consistency. It completely eliminates the biological BAC probe production process, which may cause lot-to-lot variation. |
  |
