|
A complete workflow, from DNA to result
The GenetiSure Dx Postnatal Assay uses Agilent's proprietary aCGH for copy-number and LOH data, enabling cytogeneticists to accurately detect genetic anomalies associated with developmental delay, intellectual disability, congenital anomalies, and dysmorphic features in children and adults. The GenetiSure Dx Postnatal solution includes all components required to process your microarray samples and perform data analysis.
|
|
Assay Selection
|
|

|
GenetiSure Dx Postnatal Array
- CGH + SNP microarray designed and validated for postnatal samples
- Increased resolution in targeted genomic regions of known dose-sensitivity and significant clinical interest
- Whole genome CN and LOH in a single assay
- High fidelity 60-mer oligo based on Agilent OLS technology
|
|
|
Sample Processing
|
|

|
Dx Reagents
- Labeling, Hybridization and washing reagents are validated for diagnostic and optimized for use with GenetiSure Dx Array
- Dedicated reagents allow streamlined sample processing
- Only 500 ng of genomic DNA are required for testing, extracted from 200 μl of whole blood.
- The accuracy of GenetiSure Dx Postnatal Assay results is not affected by increased levels of blood contaminates or storage for up to 7 days
|
|
|
Data Generation & Analysis
|
|

|
SureScan Dx
The SureScan Dx Microarray Scanner is the foundation of our GenetiSure Dx Postnatal Assay that offers outstanding sensitivity and resolution, delivering flexibility to analyze genomics and cytogenetics microarrays.
-> Learn more about SureScan
CytoDx
CytoDx software has been designed specifically for use with the GenetiSure Dx Postnatal Assay. The software addresses the needs of cytogeneticists for analysis and triage of their data:
- Streamlined worfklow for CN and LOH data analysis
- Validated optimized algorithms for analysis of the Agilent GenetiSure Dx Postnatal enabling suppression, classification, editing, annotations of aberrations and report generation
- Pre-loaded tracks and links to external databases for data interpreation evidence
- Software is included in our Assay, register here to download
|
|
|
|
|
Clinical Performances
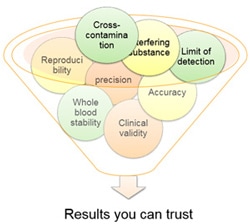
Results You Can Trust
The Genetisure Dx Postnatal assay has been validated with a Clinical study involving different partner laboratories, and including more than 900 samples.
During the study we obtained a diagnostic yield of 15% and 92.9% agreement with the reference diagnosis for Copy Number. Read more
|
|
Validation Studies
Performed on the Assay
|
|
For more information, please visit www.agilent.com/chem/contactus.
|
INTENDED USE: GenetiSure Dx Postnatal Assay is a qualitative assay intended for the postnatal detection of copy-number variations (CNV) and copy-neutral loss of heterozygosity (cnLOH) in genomic DNA obtained from peripheral whole blood in patients referred for chromosomal testing based on clinical presentation. GenetiSure Dx Postnatal Assay is intended for the detection of CNVs and cnLOH associated with developmental delay, intellectual disability, congenital anomalies or dysmorphic features. Assay results are intended to be used in conjunction with other clinical and diagnostic findings, consistent with professional standards of practice, including confirmation by alternative methods, parental evaluation, clinical genetic evaluation and counseling, as appropriate. Interpretation of assay results is intended to be performed only by healthcare professionals who are board-certified in clinical cytogenetics or molecular genetics.
The assay is intended to be used on the SureScan Dx Microarray Scanner System and analyzed by CytoDx Software.
WARNING: This device is not intended to be used for standalone or diagnostic purposes, pre-implantation or prenatal testing or screening, population screening or the detection of, or screening for, acquired or somatic genetic aberrations.
The products referenced on this page are not available for sale in all countries or jurisdictions. The information contained in this website may not be valid in your jurisdictions. Please contact your local sales representative for additional information.





