RNA Sample Quality Control Analysis
What is RNA?
Analysis of Ribonucleic acid (RNA), one of the most studied biological macromolecules, is essential in the understanding of gene expression and the regulation of cellular processes, such as cell division and cell growth. Because of these crucial roles, RNA is a molecule of interest to researchers seeking to a deeper understanding of biology. RNA must be extracted from living cells, and purified before subsequence analysis.
RNA can be found in many different subtypes, lengths and shapes, and is easily degraded. Therefore, to confidently produce robust results from RNA analyses, sample quality control (QC) ahead of downstream analysis is highly beneficial.
Agilent offers a range of quantification, qualification, and sizing solutions to allow for reliable assessment of samples. This includes challenging samples such as formalin-fixed paraffin-embedded (FFPE) RNA specimens.
For Research Use Only. Not for use in diagnostic procedures.
Total RNA Qualification
To aid in the empirical assessment of total RNA, each system features a quality metric for objective assessment of RNA quality: the RNA Integrity Number (RIN) from the Bioanalyzer system, RNA Quality Number (RQN) from the Fragment Analyzer and Femto Pulse systems, and RNA Integrity Number Equivalent (RINe) from the TapeStation systems.
Agilent solutions aid researchers by automatically calculating the quality score for every sample, taking into account ratio between ribosomal peaks, small RNA region, and resolution between peaks. The metrics are reported on a scale of 1 to 10, with the higher values indicating a higher-quality total RNA sample.
Rapid Evaluations with the 2100 Bioanalyzer System
The 2100 Bioanalyzer system offers two kits for accurate RNA Quality Control:
- The RNA 6000 Nano kit (p/n 5067-1511)
- The RNA 6000 Pico kit (p/n 5067-1513)
Both kits have a specific assay for prokaryotic and eukaryotic RNA samples and plant RNA. Features like automatic RIN evaluation, calculation of ribosomal ratios, and quantitative data offered by these assays have made the 2100 Bioanalyzer a standard for RNA QC.
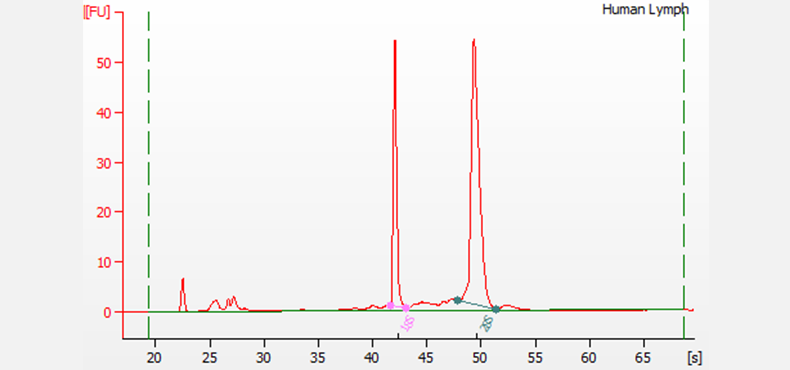
Separation of total RNA extracted from human lymph tissue. The pink and green lines designate the 18S and 28S ribosomal RNA peaks, respectively. The small fragments that appear before the ribosomal peaks are the indicative of the small RNA region. Electrophoresis was performed using the RNA 6000 Nano kit on a 2100 Bioanalyzer system. This sample has a RIN of 9.8, indicating high-quality RNA.
Consistent Fragment Analyzer Solutions for Total RNA
The Fragment Analyzer systems have two reagent kits for the consistent assessment of total RNA quality, quantity, and size:
- The RNA kit (15 nt) (p/n DNF-471)
- The HS RNA kit (15 nt) (p/n DNF-472)
ProSize data analysis software automatically calculates a quantitative quality score, the RQN, for each sample. The excellent resolution of these kits provides researchers with a clear view of the unique profiles of total RNA, separate from small and degraded RNA, making the RQN accurate.
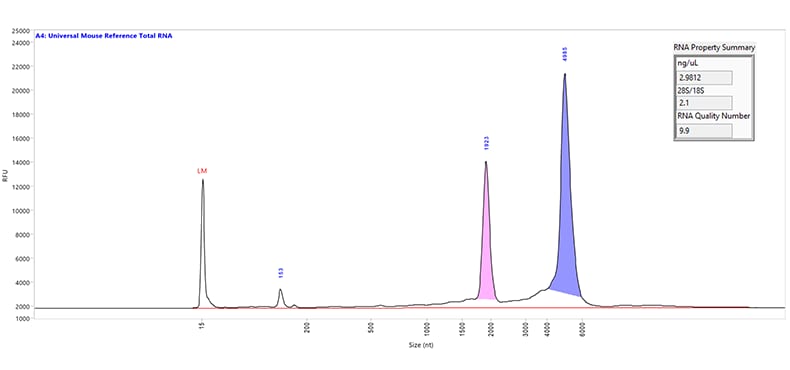
Separation of Universal Mouse Reference total RNA. The purple and pink peaks represent the large and small ribosomal RNA peaks, respectively. The small fragments seen below 200 nt is indicative of the small RNA region. Capillary electrophoresis was performed using the RNA kit (15 nt) on a Fragment Analyzer system equipped with a short capillary array, 33 cm. The RNA Property Summary (inset) includes the sample concentration, ribosomal ratio, and the RNA Quality Number (RQN). This sample has an RQN of 9.9, indicating high-quality RNA.
Efficient, Simple DNA and RNA sample QC with TapeStation Systems
The TapeStation systems offer two assays for efficient and reliable RNA characterization, quality assessment, and quantification:
- The RNA ScreenTape assay (p/n 5067–5576)
- The High Sensitivity RNA ScreenTape assay (p/n 5067–5579)
The RINe is directly comparable to the RIN of the Bioanalyzer system and delivers fast and objective evaluations of RNA integrity for both eukaryotic and prokaryotic total RNA samples.
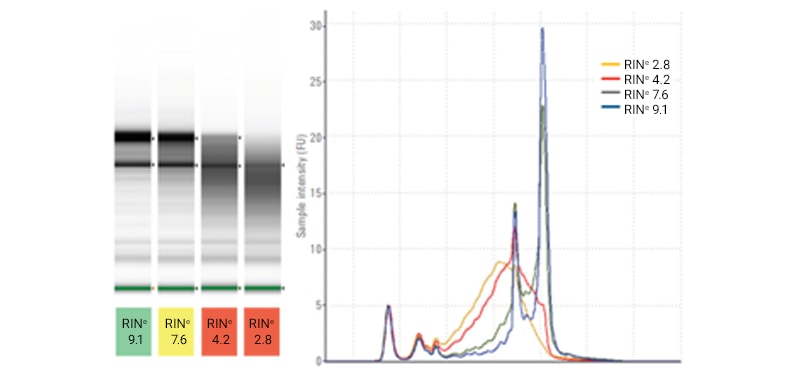
Four rat total RNA samples (300 ng/μL) with different levels of RNA degradation were analyzed on a TapeStation system using the RNA ScreenTape assay. The determined RINe values are shown under the gel image. A lower RINe indicates a more degraded sample, while a larger RINe indicates high-quality RNA.
Sensitive Femto Pulse systems
The Ultra Sensitivity RNA kit (p/n FP-1201) conserves precious samples, while properly accounting for low concentration transcripts within a total RNA sample. It enables quantitative and qualitative analysis of total RNA and mRNA from 200 to 6,000 nt at ultralow concentrations. The ProSize data analysis software and the RQN quality metric are compatible with the Femto Pulse, enabling advanced quality analysis of low concentration total RNA samples.
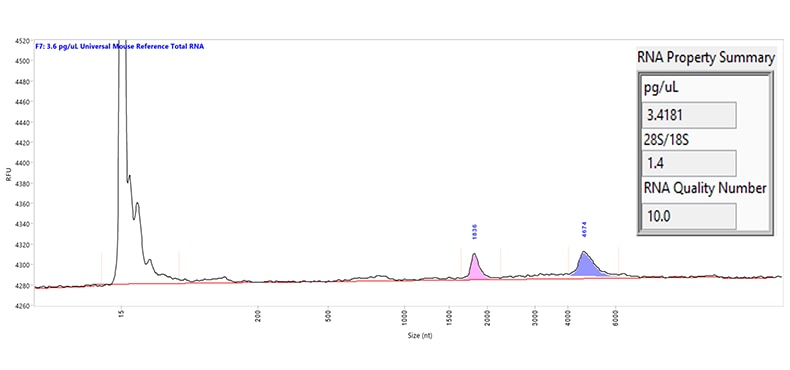
Separation of Universal Mouse Reference total RNA. The purple and pink peaks represent the large and small ribosomal RNA peaks, respectively. Capillary electrophoresis was performed using the US RNA kit on a Femto Pulse system. The RNA Property Summary (inset) includes the sample concentration, ribosomal ratio, and the RNA Quality Number (RQN). This sample has an RQN of 10.0, indicating high-quality RNA.
For Research Use Only. Not for use in diagnostic procedures.
Reduce Contamination and Misinterpretation in Messenger RNA
Messenger RNA (mRNA) provides researchers with insight into gene activity. Large-scale sequencing of mRNA enables researchers to profile what genes and genomic regions are active. In a typical workflow involving mRNA, total RNA is first analyzed for sample integrity before ribosomal RNA (rRNA) depletion, to ensure that the RNA is not degraded.
The mRNA is then analyzed with an assay to verify that the residual rRNA content is low, as indicated by a low percent rRNA contamination score. As with other NGS applications, proper mRNA QC analysis of nucleic acids ensures that only suitable samples are processed.
Quantitative applications for gene expression also rely on having intact mRNA, as degraded total RNA may lead to the absence of some transcripts, and misinterpretation of results. Sample quality assessment helps ensure the integrity of the mRNA of interest, and guarantees the sample contains minimal contaminating rRNA, which could interfere with robust downstream analysis.
Reliable QC analysis of mRNA and ribo-depleted total RNA is achieved with the Agilent Automated Electrophoresis portfolio.
For accurate assessment of mRNA, use:
- The mRNA assay for the RNA 6000 Nano (p/n 5067–1511) and RNA 6000 Pico (p/n 5067–1513) kits for the 2100 Bioanalyzer system
- The HS RNA kit (15 nt) (p/n DNF-471) for the Fragment Analyzer systems
- The Ultra Sensitivity RNA kit (p/n FP-1201) for the Femto Pulse system
2100 Bioanalyzer system
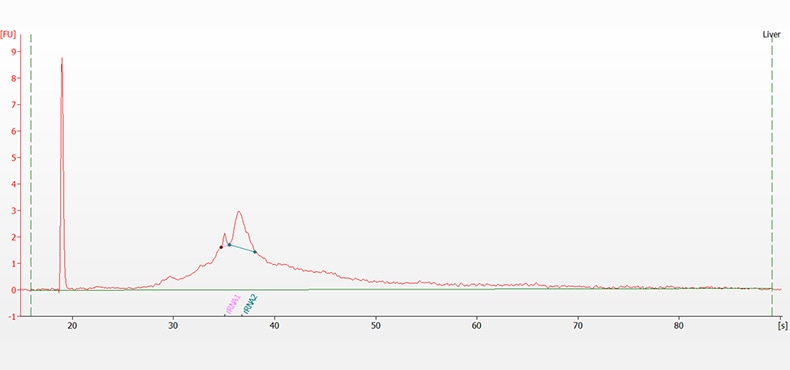
Messenger RNA (mRNA) sample extracted and purified from liver tissue. Electrophoresis was performed with the RNA 6000 Nano kit on a 2100 Bioanalyzer system using the mRNA assay. In this example, the ribosomal contamination is 8.1%.
Fragment Analyzer system
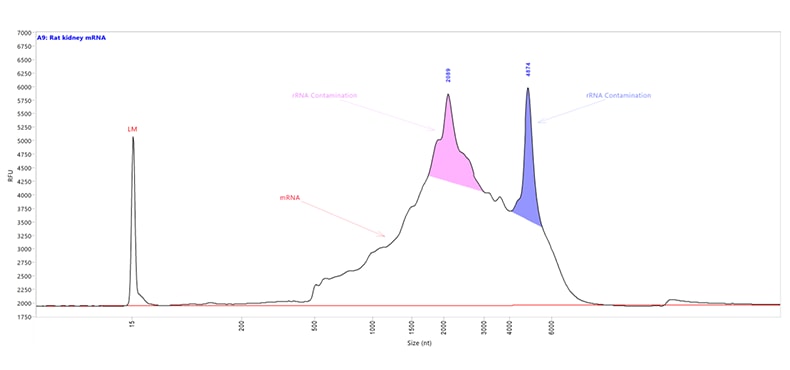
Messenger RNA (mRNA) sample extracted and purified from rat kidney tissue. Capillary electrophoresis was performed with the HS RNA kit using the mRNA separation method on a Fragment Analyzer system equipped with a short capillary array, 33 cm. The mRNA Property Summary indicates the percent of ribosomal contamination present in the sample, in this example 13.6%.
Femto Pulse system
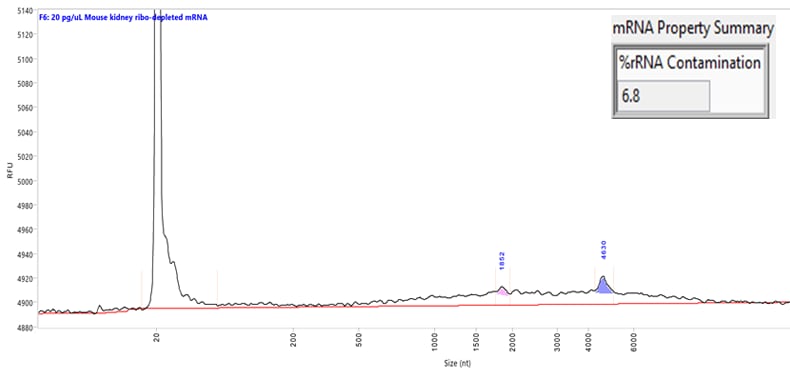
Separation of a mouse kidney mRNA sample at a low concentration (30 pg/µL). Capillary electrophoresis was performed using the Ultra Sensitivity RNA kit on a Femto Pulse system. The mRNA Property Summary indicates the percent of ribosomal contamination present in the sample, in this example 6.8%
In vitro transcribed (IVT) messenger RNA (mRNA)
in vitro transcription (IVT) mRNA is becoming widespread in research areas such as ribozyme and aptamer synthesis, mRNA synthesis, RNA interference, and antisense RNA techniques.
While conventional IVT mRNA is under 100 nucleotides in length, longer RNA transcripts are now being used for gene structure and functional studies. Reliable RNA quality control steps are required to produce these synthetic RNAs. Sizing, quantification, and quality assessment of IVT mRNA ranging from 200 to 6,000 nt in size can be obtained using the Fragment Analyzer systems with the RNA kit (15 nt) (p/n DNF-471). For longer transcripts over 6,000 nt, an extended method can be used.
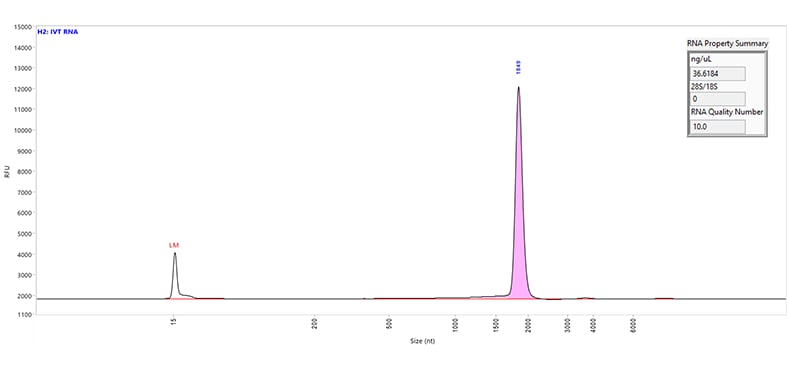
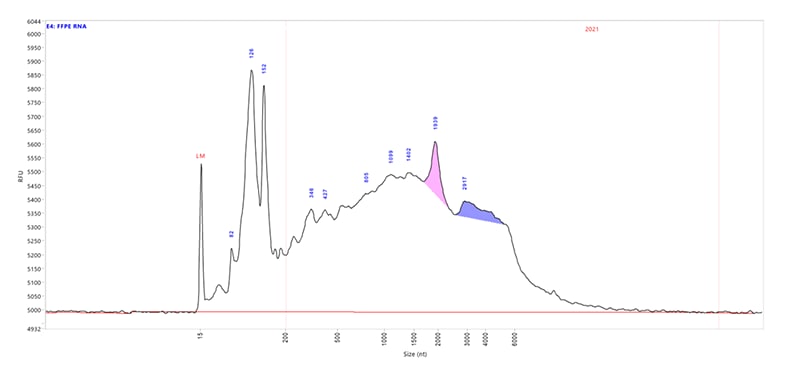
An 1,800 nt IVT mRNA sample separated using the RNA kit (15 nt) on a Fragment Analyzer system equipped with a short capillary array, 33 cm.
For Research Use Only. Not for use in diagnostic procedures.
Clear Resolution and High Sensitivity Solutions for Small RNA Analysis
Small RNA is a broad and growing classification, including: microRNA (miRNA), small interfering RNA (siRNA), PIWI-interacting RNA (piRNA), and small nuclear RNA (snRNA). Analysis of small RNAs is often difficult as many methods of analysis lack the minimum resolution and sensitivity required for accurate separation, sizing, and quantification.
The Bioanalyzer and Fragment Analyzer systems are uniquely suited for the supreme sensitivity, variable throughput, and remarkable resolution required for small RNA QC analysis with:
- The Bioanalyzer Small RNA kit (p/n 5067–1548)
- The Fragment Analyzer Small RNA kit (p/n DNF-470)
2100 Bioanalyzer system
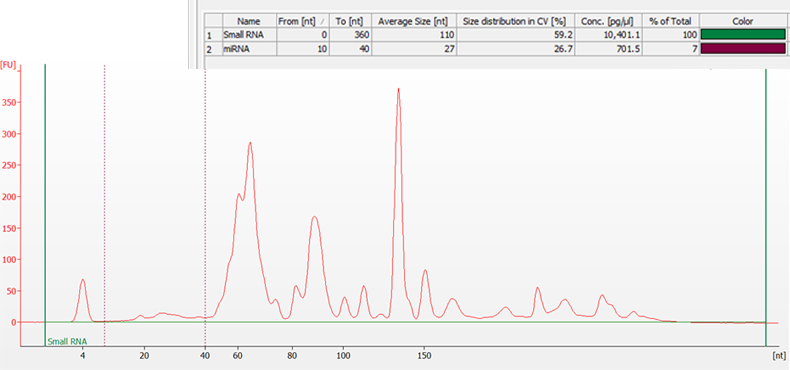
Total RNA sample extracted from mouse thymus and separated on a 2100 Bioanalyzer system using the Small RNA assay. The area between the green lines represents the Small RNA Region, while the purple dashed lines indicate the microRNA Region (10 to 40 nt). The Region Table (inset) gives the average size, size distribution, and concentration of each region, along with the percentage of the sample that is small RNA and microRNA.
Fragment Analyzer system
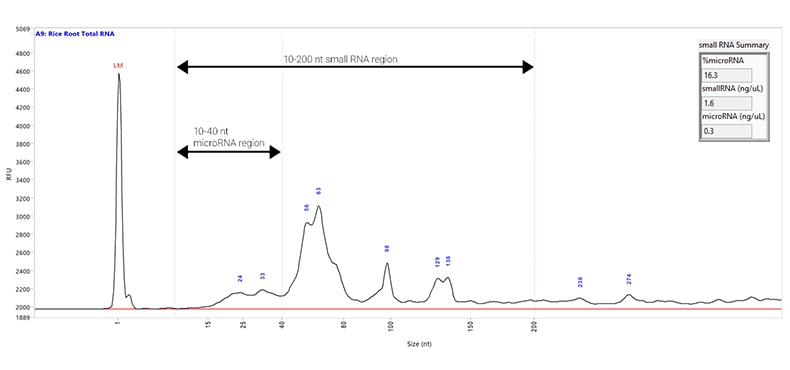
Total RNA sample extracted from rice root and separated using the Small RNA kit. Capillary electrophoresis was performed with a Fragment Analyzer system equipped with a short capillary array, 33 cm. The purple lines on the electropherogram indicate the different regions of the sample: the microRNA Region (10 to 40 nt) and the Small RNA Region (10 to 200 nt). The Small RNA Summary table (inset) gives the percentage of the sample that is microRNA, along with the concentrations of the small RNA and microRNA regions.
For Research Use Only. Not for use in diagnostic procedures.
Quality Metrics for the Unique QC Challenges of FFPE RNA
Reliable QC analysis of FFPE RNA is essential for confident NGS results. FFPE RNA samples exhibit higher levels of degradation and cross-linking, limiting the effectiveness of the RNA quality metrics for these samples.
The Automated Electrophoresis systems from Agilent utilize a quality metric, DV200, to address the unique QC challenges of FFPE RNA. The DV200 provides researchers with a tool to reliably classify degraded RNA by size and effectively parse samples suitable for NGS from unsuitable samples, conserving time and costs. Depending on the level of degradation and the DV200 score, researchers can modify their workflows, to still enable the use of FFPE samples in downstream applications. For example, when a specific size is needed and the sample is slightly degraded, the shearing protocol can be adjusted to reach that size. Take advantage of automated evaluation of RNA samples using the DV200 quality metric with any of the Automated Electrophoresis platforms:
- The Bioanalyzer RNA 6000 Nano kit (p/n 5067–1511) and RNA 6000 Pico kit (p/n 5067–1513) with the DV200 assays
- The Fragment Analyzer RNA kit (15 nt) (p/n DNF-471) and HS RNA kit (15 nt) (p/n DNF-472)
- The TapeStation RNA ScreenTape assay (p/n 5067–5576) and High Sensitivity RNA (p/n 5067–5579) ScreenTape assay
2100 Bioanalyzer system
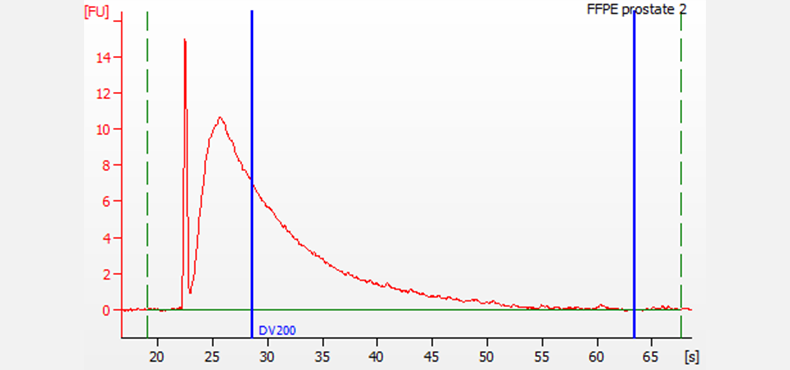
FFPE RNA extracted and purified from prostate tissue. Electrophoresis was performed using the DV200 RNA Nano assay on a 2100 Bioanalyzer system with the 2100 Expert Software. For this sample, the DV200 score is 45%.
Fragment Analyzer system
FFPE RNA sample separated using the HS RNA kit (15 nt) for the Fragment Analyzer systems. Capillary electrophoresis was performed with a short capillary array, 33 cm. In ProSize Data Analysis software, the red lines on the electropherogram indicate the DV200 region. For this sample, the DV200 score is 22.6% indicating that a small percentage of the sample is above 200 nt.
TapeStation system
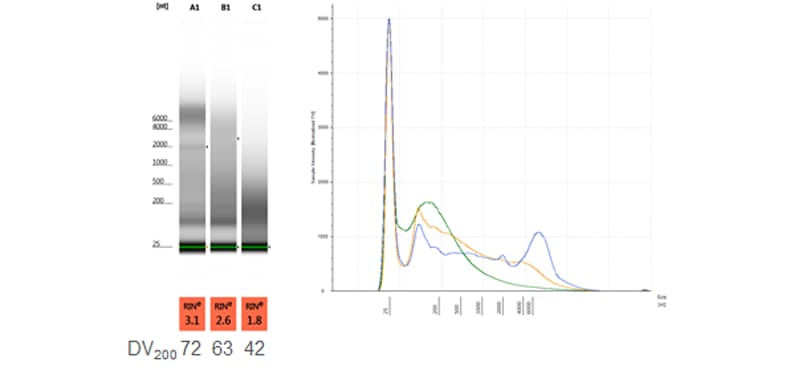
Comparison of FFPE RNA samples with varying DV200 values and typical electropherogram profiles. Samples were analyzed on a TapeStation system with the RNA ScreenTape assay.
For Research Use Only. Not for use in diagnostic procedures.
RNA Analysis Kits
The Automated Electrophoresis instruments from Agilent provide effective solutions for the QC analysis of RNA: the Bioanalyzer, Fragment Analyzer, TapeStation, and Femto Pulse systems. With the relevant reagent kits, the instruments provide unmatched sizing and quantification capabilities.
| Instrument | Part Number(s) | Reagent Kit Name | Sizing Range | Qualitative Range | Quality Metric |
|---|---|---|---|---|---|
| 2100 Bioanalyzer system | 5067–1511 | RNA 6000 Nano kit | - | 25 - 500 ng/µL | RIN, DV200 |
| 5067–1513 | RNA 6000 Pico kit | - | 50 - 5,000 pg/µl | RIN, DV200 | |
| 5067–1548 | Small RNA kit | 6 - 150 nt | 50 - 2,000 pg/µl | - | |
| Fragment Analyzer systems | DNF-470-0275 | Small RNA kit | 15 – 200 nt | 25 – 2,500 pg/µL | RQN, DV200 |
| DNF-471-0500 DNF-471-1000 |
RNA kit (15NT) | 200 – 6,000 nt | 5 – 500 ng/µL | RQN, DV200 | |
| DNF-472-0500 DNF-472-1000 |
HS RNA kit (15NT) | 200 - 6,000 nt | 50 - 5,000 pg/µL | - | |
| TapeStation systems | 5067-5576 (ScreenTape) 5067-5577 (Buffer) 5067-5578 (Ladder) |
RNA ScreenTape | - | 25 – 500 ng/μl | RINe, DV200 |
| 5067-5579 (ScreenTape) 5067-5580 (Buffer) 5067-5581 (Ladder) |
High Sensitivity RNA ScreenTape | - | 1000 – 25000 pg/μl | RINe, DV200 | |
| Femto Pulse system | FP-1201-0275 | US RNA kit | 200 – 6,000 nt | 2.5 – 250 pg/µL | RQN |
For Research Use Only. Not for use in diagnostic procedures.
Read more about RNA analysis in our application notes.
Quality Analysis of Eukaryotic Total RNA with the Agilent 5200 Fragment Analyzer SystemThe Agilent 5200 Fragment Analyzer and associated RNA Kits allow for easy analysis of total RNA quality and concentration.
Plant RNA Degradation Detection Using the Agilent 5200 Fragment Analyzer SystemThe Agilent Fragment Analyzer and associated RNA Kits allow for easy analysis of RNA quality in RT-PCR, microarray analysis, and next-generation sequencing.
Monitoring Library Preparation for Next-Generation Sequencing in systems Biology Omics AnalysisEvaluation of the Agilent 4150 TapeStation system with ScreenTape assay to monitor exome and transcriptome library preparation for systems biology omics analysis.
Comparison of RIN and RINe Algorithms for the Agilent 2100 Bioanalyzer and the Agilent 2200 TapeStation systemsThis Technical Overview demonstrates that RINe values obtained from the Agilent 2200 TapeStation system
Comparison of RIN and RQN for the Agilent Bioanalyzer and the Fragment Analyzer SystemsThis technical overview examines the RIN from the Bioanalyzer and the RQN from the Fragment Analyzer. Comparison of the RIN and RQN is performed with several eukaryotic and prokaryotic RNA samples that have various degrees of degradation.
Assessing Integrity of Insect RNAThis App Note shows the applicability of total RNA quality assessment of different insect species with the 2100 Bioanalyzer System.
Analysis of miRNA content in total RNA preparations using the Agilent 2100 BioanalyzerAdvances in miRNA research have led to the increased demand for techniques appropriate for the analysis of this small RNA subclass.
Simplified DV200 Evaluation with the Agilent 2100 Bioanalyzer SystemThis Technical Overview describes the simplified evaluation of the DV200 using the DV200 RNA Nano and DV200 RNA Pico assays with the Agilent 2100 Bioanalyzer system.
DV200 Evaluation with RNA ScreenTape AssaysThis Technical Overview describes the simplified evaluation of the DV200 using RNA ScreenTapeand High Sensitivity RNA ScreenTape on the 4200 TapeStation System.
For Research Use Only. Not for use in diagnostic procedures.