Access Agilent eNewsletter February 2015
>> Update My Profile | Subscribe to Access Agilent | Article Directory

DNA Integrity Number provides objective evidence to eliminate poor-quality sequencing samples
By Dr. Eva Schmidt
Agilent Product Manager, DNA Integrity Number (DIN)
Formalin-fixed paraffin-embedded (FFPE) tissue is widely used for clinical sample preservation and archiving, so it provides a valuable resource for Next Generation Sequencing (NGS) studies. DNA extraction from FFPE samples is challenging and often results in low amounts of suitable DNA for NGS. These DNA samples are subjected to a time-intensive NGS sample preparation workflow and only the final sequencing step reveals if meaningful results are obtained. To save time, the Agilent 2200 TapeStation assesses quality of the initial genomic DNA (gDNA), so you can objectively exclude poor-quality samples from further processing and sequencing. This article shows how a pharmaceutical laboratory has enjoyed success with this method.
Theragen Etex is a specialized genomics company developing innovative diagnosis tools and new drugs using genomics and bioinformatics technologies. The company is currently undertaking a Next Generation Sequencing project from FFPE cancer tissue samples using the Agilent SureSelectXT system for library generation and the Agilent 2200 TapeStation system for quality control within this workflow.
Replace subjective evaluation with numerical quality assessment
Agilent gave the team in Korea (shown in Figure 1) early access to a new feature for the Genomic DNA ScreenTape assay – the DNA Integrity Number (DIN) – to asses if DIN would provide a suitable quality criterion for gDNA samples extracted from FFPE tissue. This new feature is available with the latest version (revision A.01.05) of the Agilent 2200 TapeStation software. In addition to the previously available sizing and quantity information for genomic DNA samples, you can now use DIN, which ranges from 1 to 10, as a numerical assessment of the gDNA integrity. A high DIN indicates highly intact gDNA and a low DIN signifies strongly degraded gDNA.
 Enlarge
Enlarge
Figure 1. The Theragen ETEX team uses Agilent instrumentation and software to accelerate NGS library preparation.
 Enlarge
Enlarge
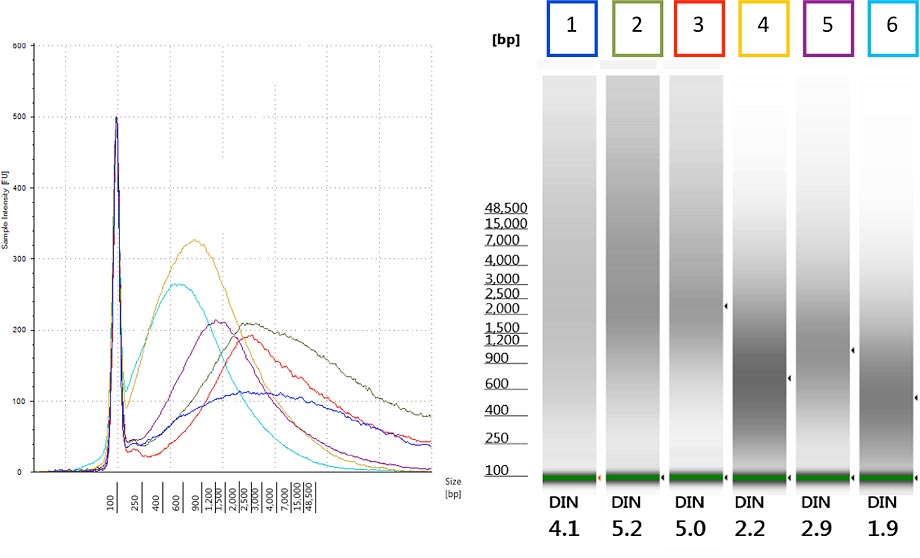
Figure 2. The Agilent DNA integrity number (DIN) provides an objective quality measure for gDNA extracted from FFPE tissue.
 Enlarge
Enlarge
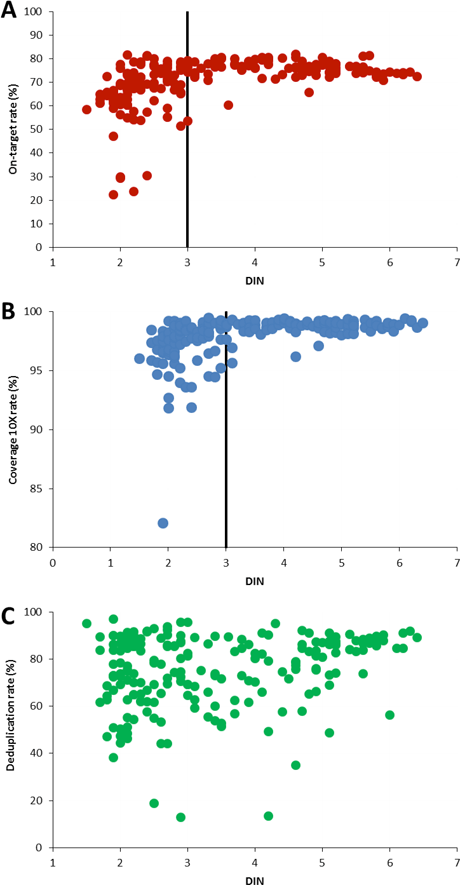
Figure 3. DIN determined with the 2200 TapeStation system and the Genomic DNA ScreenTape assay was plotted against on-target rate (A), the coverage rate at 10x (B), and the deduplication rate (C) for 197 gDNA samples extracted from FFPE cancer tissue.
The Korean researchers initially had assessed gDNA quality by subjective examination of the size distribution of samples run on the Genomic DNA ScreenTape assay. By simply upgrading the 2200 TapeStation Analysis software to revision A.01.05, they could reanalyze this historic data with the DIN algorithm. Figure 2 illustrates how difficult it can be to subjectively assess the electropherogram traces (left) or gel images (right) of the gDNA sample. However, the new software shows DIN under the gel image and in the data table, which replaces this subjectivity with a numeric measure.
DIN correlates with quality of sequencing results
A study of 751 samples of gDNA extracted from FFPE cancer tissue proved the direct correlation between DIN and the quality of downstream sequencing results. The gDNA was analyzed using the Agilent Genomic DNA ScreenTape assay and DIN ranged from 1 to 7.8. A randomly selected subset of the samples with DIN from 1.5 to 6.4 was further subjected to the Agilent SureSelectXT workflow. An on-target rate above 70% was defined as the sequencing success parameter. The coverage at 10x (target above 90%) and deduplication rate (target above 80%) provided additional quality criteria.
As part of the NGS workflow, input gDNA is fragmented, so you might assume that the original size distribution of a sample has no significant effect on the quality of the sequencing results. But our data clearly indicates the opposite. The obtained DIN was directly correlated with the on-target rate – the main success parameter of the sequencing results (Figure 3A). About one-third of the sample subset did not meet the on-target rate criterion of at least 70%. Among these, the integrity analysis showed that 97% of the samples had a DIN value below 3.
The majority of samples did pass the 90% target coverage rate at 10x, but samples with DIN below 3 showed greater variation (Figure 3B). In contrast, there was no clear correlation between DIN and the deduplication rate (Figure 3C).
A nearly 50% reduction in the number of samples processed for NGS represents a substantial saving in both time and money – and a significant acceleration in delivery of the NGS project.
Substantial reduction in workload
Based on the analysis of the initial sample integrity and the correlation to the on-target and coverage 10x rate, a DIN threshold of ≥3 was set for subsequent sequencing. Among the data subset (n=197) 49% did not pass the DIN≥3 criterion. Samples with a DIN below 3 were not further processed and sequenced, which reduced the workload by half.
Reduce costs: process only qualified samples
The Agilent 2200 TapeStation system with the automatically determined DIN is an optimal tool for high-throughput screening of gDNA samples extracted from FFPE tissue. DIN correlates with key sequencing quality metrics and allows objective exclusion of samples from further processing and sequencing. The NGS workflow is labor-intensive and the cost per sample is high, so the ability to use DIN to qualify samples is a great advantage.
In addition to DIN, the amount of gDNA extracted from the FFPE tissue is critical for the success of the downstream sequencing. The Agilent 2200 TapeStation system determines DIN and the total sample concentration in a single step.
If your lab must reduce costs, explore Agilent Application Note 5991-5360EN for details about this study. To learn more about the Genomic DNA ScreenTape assay and DIN, please visit our product page.
>> Update My Profile | Subscribe to Access Agilent | Article Directory
Figure 1.

The Theragen ETEX team uses Agilent instrumentation and software to accelerate NGS library preparation. Team members (from the left): Sanghoon Song, Junior Researcher; Hyunju Jung, Team Lead; Yeji Park, Associate Researcher and Sumin Ji, Associate Researcher.