Access Agilent eNewsletter April 2015
>> Update My Profile | Subscribe to Access Agilent | Article Directory

Measure genomic DNA damage with a sensitive, high-resolution Agilent 3D-DIP-Chip
By Nigel Skinner
Agilent Global Segment Marketing Manager
Genotoxins damage DNA and if the damage is not repaired, it produces diverse genetic mutations with far-reaching consequences. This damage causes many genetic aberrations that are linked to diseases such as cancer. Recent advances in genomic technologies enable the genome-wide analysis of the effects of genotoxins, including damage-induced changes in the transcriptome, proteome, and metabolome. To accelerate this research, Agilent developed a sensitive DIP-Chip method that uses DNA immunoprecipitation (DIP) and a microarray chip to measure the location and level of genotoxin-induced DNA damage throughout the genome.
Genomic technologies improve our ability to test the safety of novel compounds. [1] In addition, advances in sequencing technologies enable genome-wide mutation analyses. Sequencing of individual cancer genomes reveals the presence of mutational signatures embedded within cancer cells. [2] The signatures represent the sum of the exposure of an individual to various genotoxins during his lifetime, and the ability of the cells to repair DNA. These studies have revolutionized the way we view the impact of natural and manufactured products on the development and treatment of disease, offering great potential for personalized medicine.
 Enlarge
Enlarge
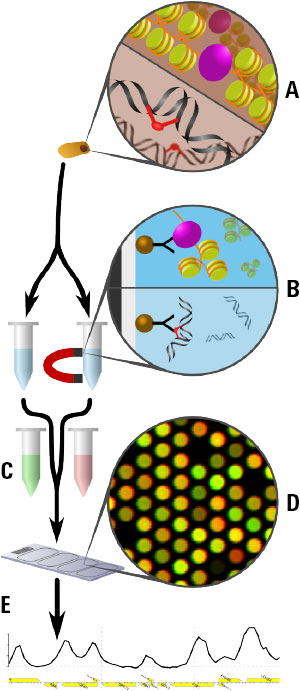
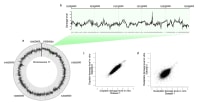
Figure 1. The Agilent 3D-DIP-Chip and ChIP-Chip procedure shown here allows you to locate DNA damage at a high resolution throughout the sequenced genome.
Optimized workflow provides detailed results
Figure 1 shows the 3D-DIP-Chip procedure. To begin, we extracted damaged DNA or cross-linked chromatin from cells (A). These were fragmented by sonication to a fragment length appropriate to the microarray. DNA fragments were captured by immunoprecipitation (IP) using magnetic beads and the appropriate antibody raised against either the specific type of DNA damage or chromatin-binding protein of interest (B).
After IP, damages or crosslinks were reversed and samples were assessed by reverse transcription polymerase chain reaction (RT-PCR) for quality control. DNA was then amplified using whole genome amplification or ligation-mediated PCR. IP samples and separate input samples were differentially labeled using the Agilent SureTag DNA Labeling Kit (C).
Samples were then hybridized to the microarray using an Agilent ChIP-on-Chip Hybridization Kit, followed by washing and drying with the Agilent ChIP-on-Chip Wash Buffer Kit (D). The microarray was then scanned as described in the Agilent mammalian ChIP-on-chip protocol using the Agilent SureScan Microarray Scanner. Finally, feature extraction software delivered data that we analyzed and plotted using the R software Sandcastle package (E). [3]
 Enlarge
Enlarge
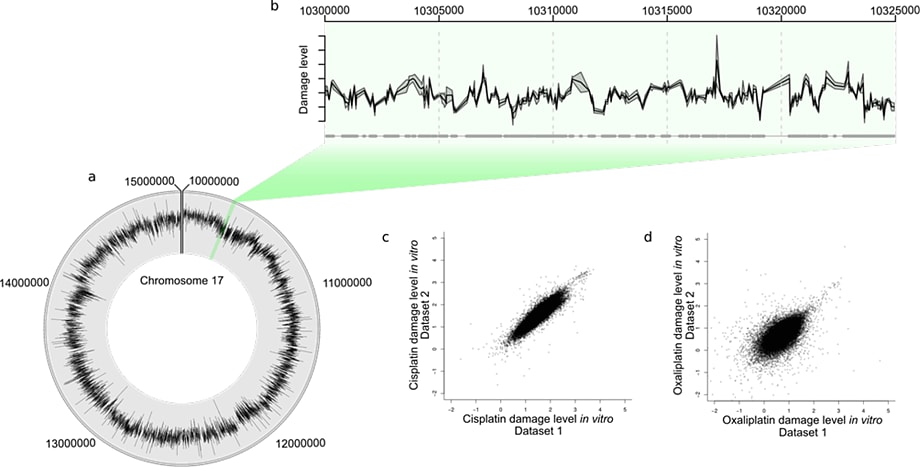
Figure 2. Human in vivo damage caused by a platinating agent. Plot D has the same axis limits as plot C and so a few probes are not shown.
 Enlarge
Enlarge
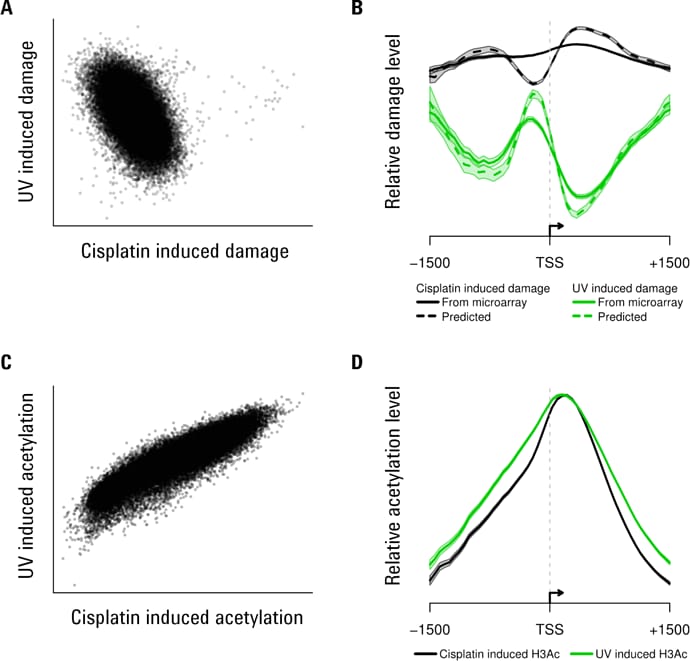
Figure 3. Some examples of Agilent 3D-DIP-Chip and ChIP-chip analyses show both positive and negative correlations.
Microarray processing and data analysis pinpoints DNA damage
We looked at human- and yeast-derived samples to determine the relative levels and locations of DNA damage. These were hybridized to two-color Agilent 4×44K microarrays, of a custom human design that covered 10 Mbp of chromosome 17, or a yeast whole-genome array. All analyses used values of log2 IP:IN, in which the input channel (IN) corrected for relative differences in DNA amounts. This procedure produced relative levels of DNA adducts detected at the genomic locations represented by the probes or features printed on the microarray.
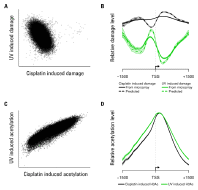
The Circos plot in Figure 2A shows the average of two in vivo human cisplatin-treated data sets. You can see the heterogeneity of the DNA damage distribution over the 10 Mbp genome section. Figure 2B shows a 25 Kbp section, with the mean (black line) and standard error (gray shading) for the two data sets. A scatterplot of one data set against the other for the total cisplatin (C) and oxaliplatin (D) data reveals the overall reproducibility.
We measured genomic DNA damage profiles for two different forms of DNA damage in human DNA: chemically (platinum) induced lesions and UV-induced pyrimidine dimers (Figures 2 and 3). Genetic damage also induces epigenetic changes in the form of histone modifications [4,5] and these changes affect induction of DNA damage and its removal by DNA repair. To investigate how these two parameters relate to each other in response to genotoxic exposure, we treated yeast cells with the same two classes of genotoxins. We also used ChIP-chip to measure DNA-damage-induced changes in histone H3 acetylation (H3Ac) at lysine 9 (K9), which is a known requirement for efficient repair of UV-induced cyclobutane pyrimidine dimers (Figure 3).
In Figure 3, scatterplots show an inverse association between cisplatin and UV-induced DNA damage (A) but a positive association between cisplatin and UV-induced histone acetylation (C). Plotting the data around transcription start sites (TSSs) shows different patterns of damage induction (B). The shaded region shows standard errors for all TSSs. Histone acetylation around the same TSSs shows similar patterns with both damaging agents (D).
Better way to measure genomic DNA damage
The Agilent DIP-Chip assay is now available for measuring various types of DNA damage in human cells with sensitivity and high resolution. This assay represents a significant technological advance in the measurement of genetic damage at a genomic scale. It now offers a novel way to examine genomic DNA damage in human and other cells. Take a moment to learn more about Agilent solutions for gene regulation.
References
- Perkel, J. M. Science, 2012, 335, 1122–1125.
- Alexandrov, L. B. Nature, 2013, 500, 415.
- R: Development Core Team. R: A Language and Environment for Statistical Computing. R: Foundation for Statistical Computing, Vienna, Austria, 2011.
- Yu, S. et al. PLoS Genetics, 2011, 7(6), e1002124.
- Guo, R. et al. Nucleic Acids Res. 2011, 39(4), 1390–1397.
>> Update My Profile | Subscribe to Access Agilent | Article Directory