Access Agilent eNewsletter, July 2013
>> Update My Profile | Subscribe to Access Agilent | Article Directory

2-D LC/MS heart-cut approach reduces method development for pharmaceutical impurity identification
By Siji Joseph
Agilent Application Scientist
Detection and identification of impurities in active pharmaceutical ingredients (APIs) are critical in the pharmaceutical industry, and LC/MS is frequently the technique of choice. Analysts often start with a method that was developed for a conventional LC detector and then convert it to an LC/MS method. The caveat is that MS is limited to the use of volatile buffers, so if your initial LC separation used nonvolatile buffers, you have a problem. One solution is to use heart-cutting to transfer the impurity peak to a second column that uses a volatile buffer for MS analysis.
Recently, we demonstrated the separation of a close-eluting impurity using heart-cut two-dimensional LC/MS. To perform the experiment, we used the Agilent 1260 Infinity Binary LC System with an additional binary pump, coupled with an Agilent 6540 Q-TOF LC/MS System.
 Enlarge
Enlarge
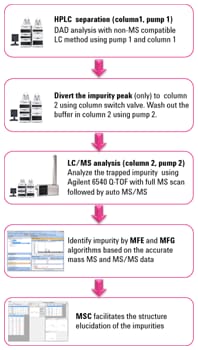
Figure 1. Workflow for impurity identification using a heart-cut approach. For MS analysis, use the same LC method you developed for a conventional LC detector.
Data processing used advanced Agilent MassHunter algorithms that allowed impurity identification and structure elucidation. These algorithms included molecular feature extraction (MFE) and molecular formula generation (MFG), along with MassHunter Molecular Structure Correlator (MSC). We demonstrated the entire workflow by identification of an impurity observed in duloxetine drug substance.
Eliminate the need to develop an MS-compatible method
We performed the impurity analysis of duloxetine API with a validated non-MS compatible liquid chromatography literature method, and an unknown impurity eluted towards the end of the main peak. We used an Agilent ZORBAX Eclipse Plus C-18, 4.6 × 250 mm, 5.0 µm column with pump 1 for the first dimension separation. A 2-position/6-port column switching valve diverted this impurity to the second LC column, where we then separated it using an MS-compatible mobile phase from the second pump.
This approach eliminated the need to develop an MS-compateed to develop an MS-compat accurate mass MS and MS/MS data of the impurity with the Agilent 6540 Q-TOF. Data processing with Agilent MassHunter Workstation and Agilent Molecular Structure Correlator (MSC) software enabled quick identification of the unknown impurity. Figure 1 shows the workflow for the experiment.
For this experiment, we used an Agilent 6540 Q-TOF LC/MS with a Jet Stream source, and an Agilent 1260 Infinity Binary LC System with the following modules and columns:
- Agilent 1260 Infinity Degasser for the first and second dimensions
- Two Agilent 1260 Infinity Binary Pumps for the first and second dimensions
- Agilent 1260 Infinity High Performance Autosampler
- Agilent 1260 Infinity Thermostatted Column Compartment with 2-position/6-port column switching valve
- Agilent 1290 Infinity Diode Array Detector with Max-Light flow cell
(4.0 µL volume, 60 mm path length) - Column 1: Agilent ZORBAX Eclipse Plus C-18, 4.6 × 250 mm, 5.0 µm
- Column 2: Agilent ZORBAX Eclipse Plus C-18, 4.6 × 75 mm, 3.5 µm
 Enlarge
Enlarge
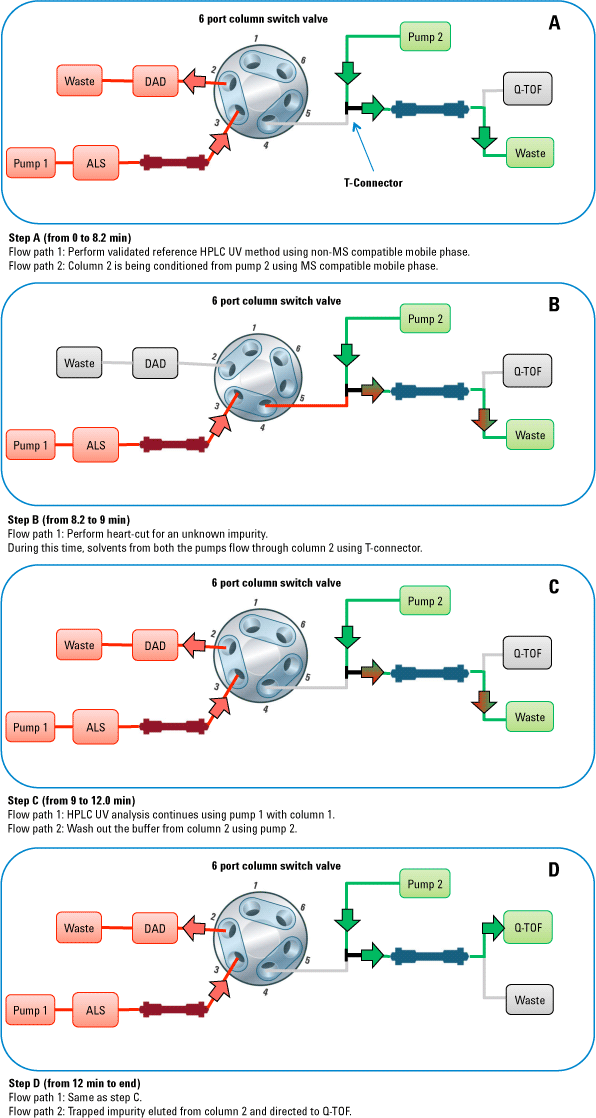
Figure 2. Valve positions before, during, and after the heart-cut process. Red indicates the flow path (1) with non-MS-compatible mobile phase and green shows the flow path (2) with MS-compatible mobile phase.
2-position/6-port column switching valve made the heart-cut easy
Figure 2 shows the valve positions during the heart-cut method. The impurity was eluted from column 1 at 8.7 minutes using method 1. The unknown impurity was then transferred to column 2 by changing the switching valve position between 8.2 min to 9.0 min. Agilent Application Note 5991-1873EN provides further details.
Use Agilent MassHunter Molecular Structure Correlator (MSC) for impurity identification
We performed data analysis using the MFE and MFG algorithms of MassHunter Qualitative Analysis. The MFE algorithm extracted high-resolution accurate mass spectra of all the sample components. From the acquired data, the MFE algorithm listed two entities, with m/z values of 298.1257 and 312.1416.
The MFG algorithm tabulated a list of candidate molecular formulas for each entity and ranked them according to their relative probabilities. We uploaded the accurate mass information of the precursor and fragment ions of both entities to the MSC software and searched against the ChemSpider database to retrieve all possible structures. The search results confirmed that m/z 298.1257 corresponded with the duloxetine API and the unknown impurity was the methyl derivative of duloxetine.
Learn how you can achieve results like these
You can find additional details and more results in Agilent Application Note 5991-1873EN. Then talk with your Agilent Representative to ensure that you have everything you need to identify unknown impurities in drug substances. And now learn all about our best in class Agilent 1290 Infinity 2D-LC solutions for doing both heart-cut and comprehensive 2D-LC applications effortlessly.
>> Update My Profile | Subscribe to Access Agilent | Article Directory